Group Analysis¶
- CPAC.group_analysis.create_fsl_flame_wf(ftest=False, wf_name='groupAnalysis')[source]¶
FSL FEAT BASED Group Analysis
- Parameters:
- ftestboolean, optional(default=False)
Ftest help investigate several contrasts at the same time for example to see whether any of them (or any combination of them) is significantly non-zero. Also, the F-test allows you to compare the contribution of each contrast to the model and decide on significant and non-significant ones
- wf_namestring
Workflow name
- Returns:
- grp_analysisworkflow object
Group Analysis workflow object
Notes
Workflow Inputs:
inputspec.mat_file : string (existing file) Mat file containing matrix for design inputspec.con_file : string (existing file) Contrast file containing contrast vectors inputspec.grp_file : string (existing file) file containing matrix specifying the groups the covariance is split into inputspec.zmap_files : string (existing nifti file) derivative or the zmap file for which the group analysis is to be run inputspec.z_threshold : float Z Statistic threshold value for cluster thresholding. It is used to determine what level of activation would be statistically significant. Increasing this will result in higher estimates of required effect. inputspec.p_threshold : float Probability threshold for cluster thresholding. inputspec.fts_file : string (existing file) file containing matrix specifying f-contrasts inputspec.paramerters : string (tuple) tuple containing which MNI and FSLDIR path information
Workflow Outputs:
outputspec.merged : string (nifti file) 4D volume file after merging all the derivative files from each specified subject. outputspec.zstats : list (nifti files) Z statistic image for each t contrast outputspec.zfstats : list (nifti files) Z statistic image for each f contrast outputspec.fstats : list (nifti files) F statistic for each contrast outputspec.cluster_threshold : list (nifti files) the thresholded Z statistic image for each t contrast outputspec.cluster_index : list (nifti files) image of clusters for each t contrast; the values in the clusters are the index numbers as used in the cluster list. outputspec.cluster_localmax_txt : list (text files) local maxima text file for each t contrast, defines the coordinates of maximum value in the cluster outputspec.overlay_threshold : list (nifti files) 3D color rendered stats overlay image for t contrast After reloading this image, use the Statistics Color Rendering GUI to reload the color look-up-table outputspec.overlay_rendered_image : list (nifti files) 2D color rendered stats overlay picture for each t contrast outputspec.cluster_threshold_zf : list (nifti files) the thresholded Z statistic image for each f contrast outputspec.cluster_index_zf : list (nifti files) image of clusters for each f contrast; the values in the clusters are the index numbers as used in the cluster list. outputspec.cluster_localmax_txt_zf : list (text files) local maxima text file for each f contrast, defines the coordinates of maximum value in the cluster outputspec.overlay_threshold_zf : list (nifti files) 3D color rendered stats overlay image for f contrast After reloading this image, use the Statistics Color Rendering GUI to reload the color look-up-table outputspec.overlay_rendered_image_zf : list (nifti files) 2D color rendered stats overlay picture for each f contrast
Order of commands:
Merge all the Z-map 3D images into 4D image file. For details see fslmerge:
fslmerge -t sub01/sca/seed1/sca_Z_FWHM_merged.nii sub02/sca/seed1/sca_Z_FWHM.nii.gz .... merge.nii.gz arguments -t : concatenate images in time
Create mask specific for analysis. For details see fslmaths:
fslmaths merged.nii.gz -abs -Tmin -bin mean_mask.nii.gz arguments -Tmin : min across time -abs : absolute value -bin : use (current image>0) to binarise
FSL FLAMEO to perform higher level analysis. For details see flameo:
flameo --copefile = merged.nii.gz --covsplitfile = anova_with_meanFD.grp --designfile = anova_with_meanFD.mat --fcontrastsfile = anova_with_meanFD.fts --ld=stats --maskfile = mean_mask.nii.gz --runmode=ols --tcontrastsfile = anova_with_meanFD.con arguments --copefile : cope regressor data file --designfile : design matrix file --maskfile : mask file --tcontrastsfile : file containing an ASCII matrix specifying the t contrasts --fcontrastsfile : file containing an ASCII matrix specifying the f contrasts --runmode : Interference to perform (mixed effects - OLS)
Run FSL Easy thresh
Easy thresh is a simple script for carrying out cluster-based thresholding and colour activation overlaying:
easythresh <raw_zstat> <brain_mask> <z_thresh> <prob_thresh> <background_image> <output_root> [--mm]
A seperate workflow called easythresh is called to run easythresh steps.
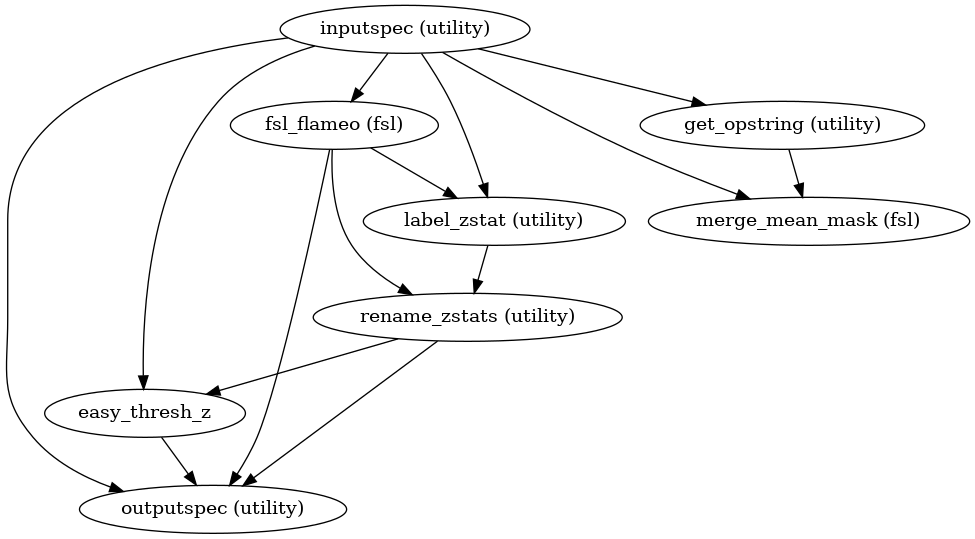
High Level Workflow Graph:
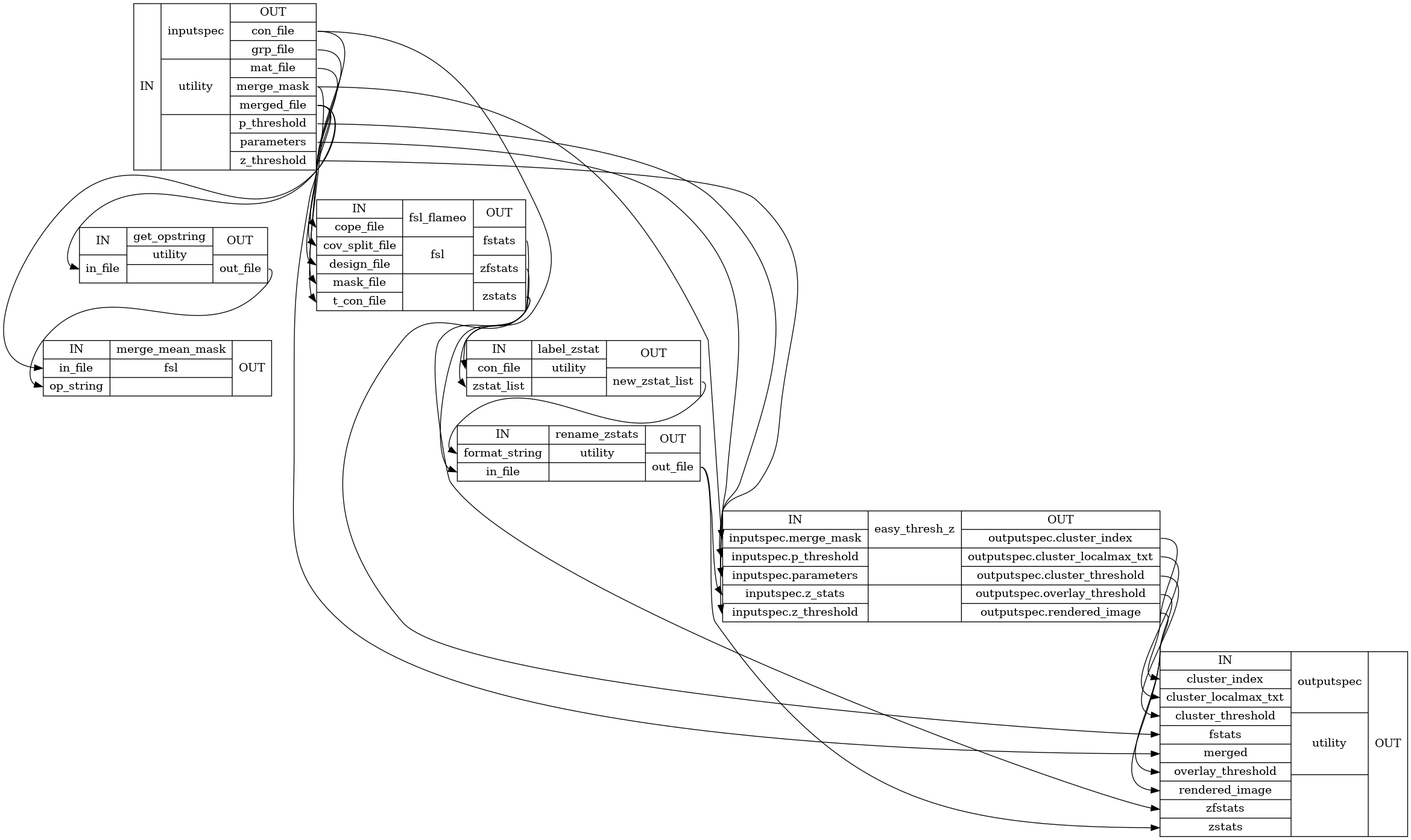
Detailed Workflow Graph:
Examples
>>> from CPAC.group_analysis import create_fsl_flame_wf >>> preproc = create_fsl_flame_wf() >>> preproc.inputs.inputspec.mat_file = '../group_models/anova_with_meanFD/anova_with_meanFD.mat' >>> preproc.inputs.inputspec.con_file = '../group_models/anova_with_meanFD/anova_with_meanFD.con' >>> preproc.inputs.inputspec.grp_file = '../group_models/anova_with_meanFD/anova_with_meanFD.grp' >>> preproc.inputs.inputspec.zmap_files = [ ... 'subjects/sub01/seeds_rest_Dickstein_DLPFC/sca_Z_FWHM.nii.gz', ... 'subjects/sub02/seeds_rest_Dickstein_DLPFC/sca_Z_FWHM.nii.gz' ... ] >>> preproc.inputs.inputspec.z_threshold = 2.3 >>> preproc.inputs.inputspec.p_threshold = 0.05 >>> preproc.inputs.inputspec.parameters = ('/usr/local/fsl/', 'MNI152') >>> preproc.run()
