Seed-based Correlation Analysis (SCA) and Dual Regression¶
Connectivity in the Brain¶
Brain connectivity can refer to multiple distinct concepts, and it is important to understand the differences between them. When referring to anatomy, connectivity may refer to physical connections between brain areas, such as the long-distance fiber tracts revealed through methods such as Diffusion Tensor Imaging. Another form of connectivity that is commonly discussed in the literature is “functional connectivity”, which is what we will be focusing on here.
What is Functional Connectivity?¶
Perhaps the most confusing thing to understand about functional connectivity is that in most cases, connectivity is never actually measured. This is because the term “functional connectivity” has come to refer to similarities in patterns of brain activity between regions. Two regions are said to be functionally connected if the time series of their activity is highly correlated. The reasoning behind this definition is that brain areas with similar activity patterns are likely to be communicating and sharing information.
Seed-based Correlation Analysis¶
Seed-based Correlation Analysis (SCA) is one of the most common ways to explore functional connectivity within the brain. Based on the time series of a seed voxel (or ROI), connectivity is calculated as the correlation of time series for all other voxels in the brain. The result of SCA is a connectivity map showing Z-scores for each voxel indicating how well its time series correlates with the time series of the seed. Below is an example connectivity map showing correlated voxels based on a seed in the precuneus.
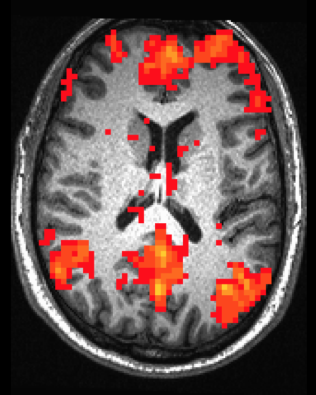
Astute readers will note that the pattern of connectivity mapped above closely resembles the anatomy of the Default Network. Indeed, SCA may be used to explore functional networks that share similar patterns of activity.
Dual Regression¶
Many large-scale functional brain networks have been shown to be reproducible across subjects (Smith et al, 2009; Yeo et al, 2011). Dual Regression (Beckman et al, 2009; Filippini et al, 2009) allows researchers to investigate individual and group differences in the structure of these networks, as well as to identify subject-specific networks based on networks identified at the group level.
The following steps are performed for each spatial map provided for Dual Regression:
Spatial Regression: The spatial map is used as a spatial regressor in a GLM to find the time series associated with the voxels in that map.
Temporal Regression: The time series found by spatial regression is then used as a temporal regressor to find the full set of voxels associated with that time series.
The result of these steps is a subject-specific spatial map based on the original spatial map.
The spatial maps used during Dual Regression can be generated any number of ways, but a common practice is to use Independent Component Analysis (Smith et al, 2009) or clustering algorithms (Yeo et al, 2011) to generate group-specific maps of large-scale brain networks.
Configuring C-PAC¶
Before SCA can be calculated, you must first extract a seed time series from which to calculate correlations. This is done by configuring and running Time Series Extraction. You will not be able to proceed in the GUI without first entering in an ROI Specification, and will receive an error if the specification is not defined.
Seed-based Correlation (SCA) - [On, Off]: For each extracted ROI Average and/or ROI Voxelwise time series, CPAC will generate a whole-brain correlation map. It should be noted that for a given seed/ROI, SCA maps for ROI Average and ROI Voxelwise time series will be the same.
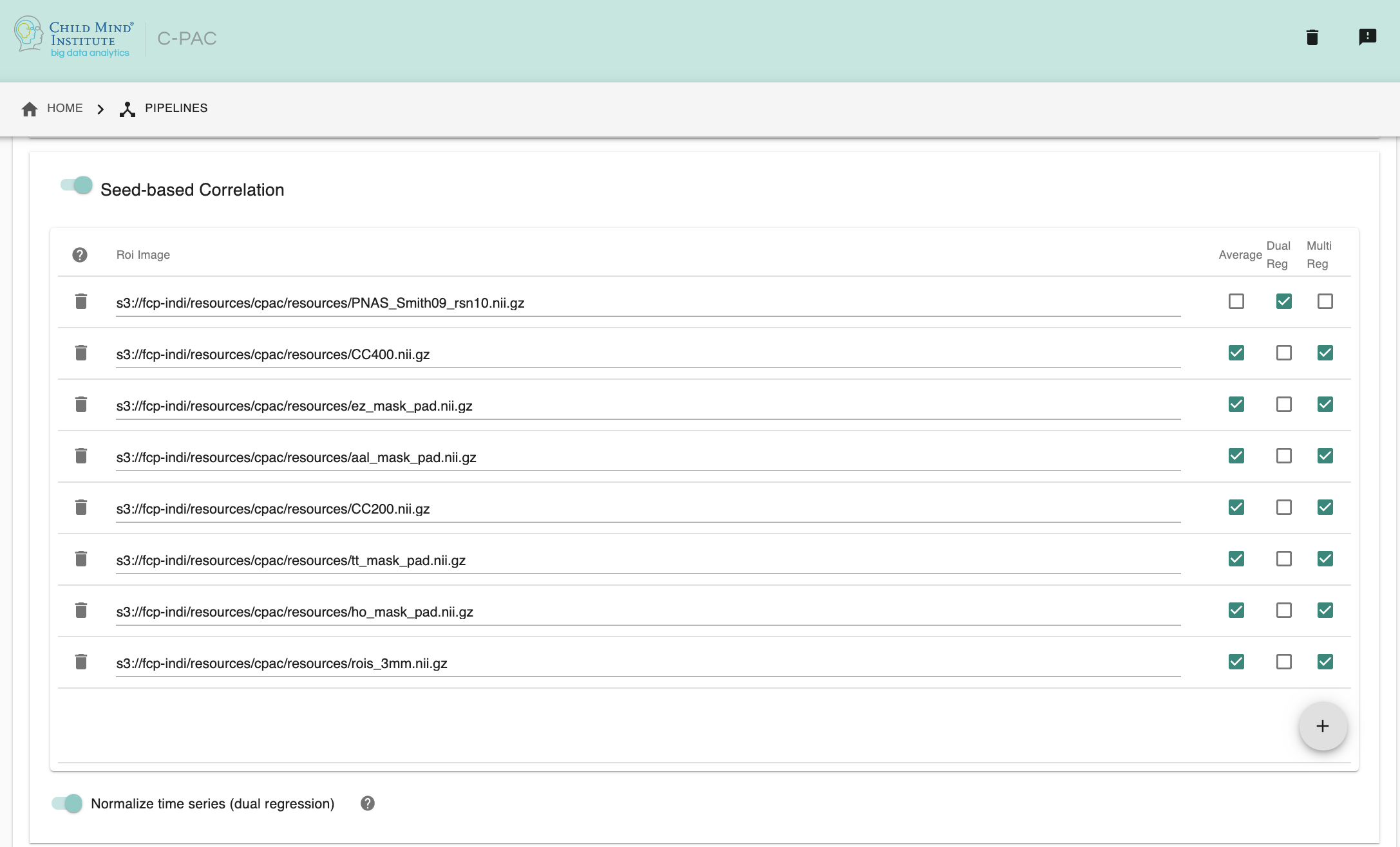
- SCA ROI Paths - [path dialogue]: Clicking on the + icon to the right of the box here will bring up a dialog where you can define multiple paths to NifTIs containing ROI masks. You may add multiple ROIs to the box. Three columns within the box can be checked on and off to enable specific options for SCA/Dual Regression:
Avg - Run seed-based correlation analysis using the average time series of each ROI.
DualReg - Run dual regression for each ROI.
MultiReg - Run a multiple regression SCA where C-PAC will enter all extracted time series from ROI Average TSE, ROI Voxelwise TSE, and Spatial Regression into a single multiple regression model and output a single correlation map.
Normalize Time Series (Dual Regression) - [On, Off]: Normalize each time series before running Dual Regression.
Configuration Without the GUI¶
The following nested key/value pairs will be set to these defaults if not defined in your pipeline configuration YAML.
Unlike most lines in a C-PAC pipeline configuration file, the keys under *_roi_paths are user-definable.
If you exclude *_roi_paths from your pipeline configuration, the default atlases and analyses will be used.
If you include *_roi_paths, only the atlases and analyses provided will be used, unless you add the optional key/value pair roi_paths_fully_specified: False in the relevant section, in which case the default atlases and analyses will be updated with the atlases and analyses specified in your pipeline config. For example,
timeseries_extraction:
roi_paths_fully_specified: False
seed_based_correlation_analysis:
roi_paths_fully_specified: False
would keep any default atlases not respecified in your pipeline configuration.
seed_based_correlation_analysis:
# SCA - Seed-Based Correlation Analysis
# For each extracted ROI Average time series, CPAC will generate a whole-brain correlation map.
# It should be noted that for a given seed/ROI, SCA maps for ROI Average time series will be the same.
run: Off
# Enter paths to region-of-interest (ROI) NIFTI files (.nii or .nii.gz) to be used for seed-based correlation analysis, and then select which types of analyses to run.
# Denote which analyses to run for each ROI path by listing the names below. For example, if you wish to run Avg and MultReg, you would enter: '/path/to/ROI.nii.gz': Avg, MultReg
# available analyses:
# /path/to/atlas.nii.gz: Avg, DualReg, MultReg
sca_roi_paths:
/cpac_templates/PNAS_Smith09_rsn10.nii.gz: DualReg
/cpac_templates/CC400.nii.gz: Avg, MultReg
/cpac_templates/ez_mask_pad.nii.gz: Avg, MultReg
/cpac_templates/aal_mask_pad.nii.gz: Avg, MultReg
/cpac_templates/CC200.nii.gz: Avg, MultReg
/cpac_templates/tt_mask_pad.nii.gz: Avg, MultReg
/cpac_templates/ho_mask_pad.nii.gz: Avg, MultReg
/cpac_templates/rois_3mm.nii.gz: Avg, MultReg
# Normalize each time series before running Dual Regression SCA.
norm_timeseries_for_DR: True
References¶
C.F. Beckmann, C.E. Mackay, N. Filippini, and S.M. Smith. Group comparison of resting-state FMRI data using multi-subject ICA and dual regression. OHBM, 2009.
Filippini, B.J. MacIntosh, M.G. Hough, G.M. Goodwin, G.B. Frisoni, S.M. Smith, P.M. Matthews, C.F. Beckmann and C.E. Mackay. Distinct patterns of brain activity in young carriers of the APOE-ε4 allele. PNAS, 106(17):7209-14, 2009.
Smith, S. M., Fox, P. T., Miller, K. L., Glahn, D. C., Fox, P. M., Mackay, C. E., et al. (2009). Correspondence of the brain’s functional architecture during activation and rest. Proceedings of the National Academy of Sciences of the United States of America, 106(31), 13040–13045. doi:10.1073/pnas.0905267106
Thomas Yeo, B. T., Krienen, F. M., Sepulcre, J., Sabuncu, M. R., Lashkari, D., Hollinshead, M., et al. (2011). The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of Neurophysiology, 106(3), 1125–1165. doi:10.1152/jn.00338.2011
