Anatomical Preprocessing¶
Surface Analysis¶
Surface analysis runs FreeSurfer recon-all and generates CSF, WM, GM masks, pial surface mesh, smoothed surface mesh, spherical surface mesh, white matter surface mesh, sulcal depth surface maps, cortical thickness surface maps and cortical volume surface maps.
Configuring CPAC to run surface analysis:¶

FreeSurfer - [On,Off]: FreeSurfer recon-all. Default is Off.
Configuration Without the GUI¶
The following nested key/value pairs will be set to these defaults if not defined in your pipeline configuration YAML.
surface_analysis:
# Will run Freesurfer for surface-based analysis. Will output traditional Freesurfer derivatives.
# If you wish to employ Freesurfer outputs for brain masking or tissue segmentation in the voxel-based pipeline,
# select those 'Freesurfer-' labeled options further below in anatomical_preproc.
freesurfer:
# If anatomical_preproc['brain_extraction']['using'] includes FreeSurfer-ABCD and this switch is On, C-PAC will automatically turn this switch Off to avoid running FreeSurfer twice unnecessarily
run: Off
# Add extra arguments to recon-all command
reconall_args: None
# (Optional) Provide an already-existing FreeSurfer output directory to ingress already-computed surfaces
freesurfer_dir: None
# Run ABCD-HCP post FreeSurfer and fMRISurface pipeline
post_freesurfer:
run: Off
subcortical_gray_labels: /opt/dcan-tools/pipeline/global/config/FreeSurferSubcorticalLabelTableLut.txt
freesurfer_labels: /opt/dcan-tools/pipeline/global/config/FreeSurferAllLut.txt
surf_atlas_dir: /opt/dcan-tools/pipeline/global/templates/standard_mesh_atlases
gray_ordinates_dir: /opt/dcan-tools/pipeline/global/templates/91282_Greyordinates
gray_ordinates_res: 2
high_res_mesh: 164
low_res_mesh: 32
fmri_res: 2
smooth_fwhm: 2
Initial Preprocessing¶
Initial preprocessing offers methods like ACPC Alignment , non-local means filtering and N4 bias field correction to preprocess anatomical images.
C-PAC provides options for configuring initial preprocessing - users can select:
Configuring CPAC to run initial preprocessing:¶
ACPC Alignment - [On,Off]: Anterior Commissure - Posterior Comissure (ACPC) alignment. Default is Off. If choose ‘on’, clicking on the setting icon will bring up a dialog where you can set ACPC alignment parameters.
Non-Local Means Filtering - [On,Off]: ANTs DenoiseImage. Default is Off.
N4 Bias Field Correction - [On,Off]: ANTs N4BiasFieldCorrection - a variant of the popular N3 (nonparametric nonuniform normalization) retrospective bias correction algorithm. Default is Off.
Configuration Without the GUI¶
The following nested key/value pairs will be set to these defaults if not defined in your pipeline configuration YAML.
anatomical_preproc:
run: On
run_t2: Off
# Non-local means filtering via ANTs DenoiseImage
non_local_means_filtering:
# this is a fork option
run: [Off]
# options: 'Gaussian' or 'Rician'
noise_model: 'Gaussian'
# N4 bias field correction via ANTs
n4_bias_field_correction:
# this is a fork option
run: [Off]
# An integer to resample the input image to save computation time. Shrink factors <= 4 are commonly used.
shrink_factor: 2
# Bias field correction based on square root of T1w * T2w
t1t2_bias_field_correction:
run: Off
BiasFieldSmoothingSigma: 5
acpc_alignment:
run: Off
Configuring ACPC Alignment options:¶
Note: These options are pre-set for ACPC Alignment’s default values. These do not need to be modified unless you are looking to optimize the results of ACPC alignment for your particular dataset.
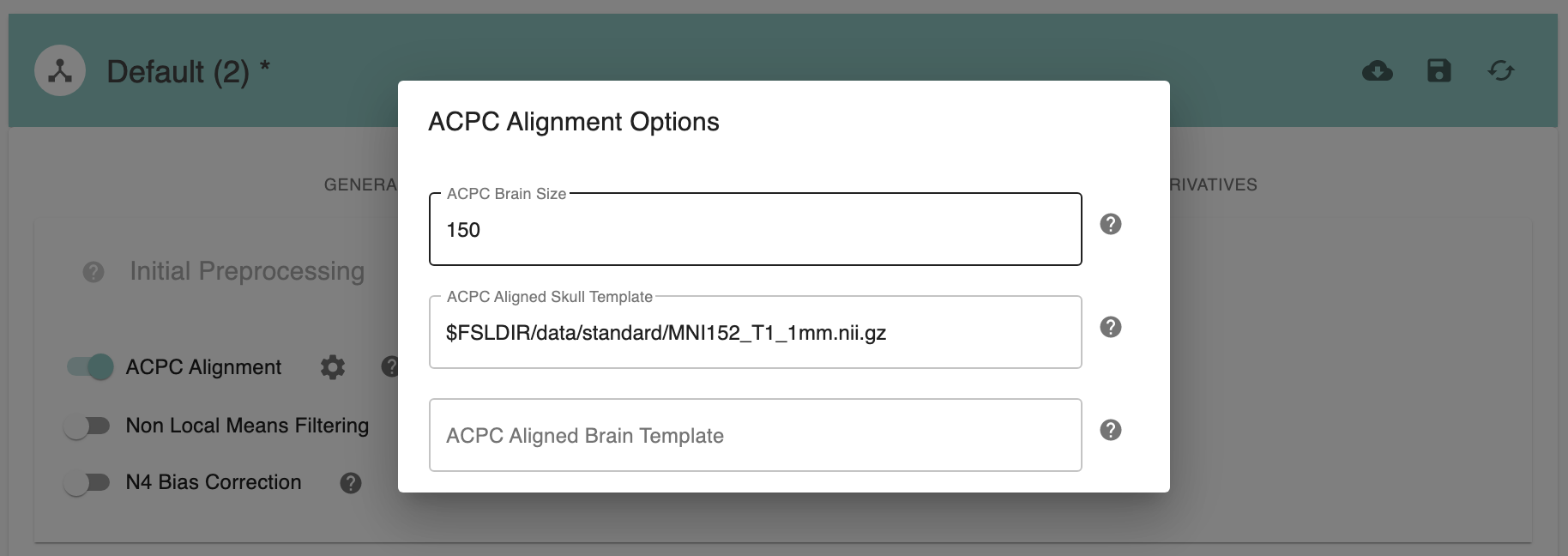
ACPC Brain Size - [150]: ACPC size of brain in z-dimension in mm. Default: 150mm for human data, 70mm for macaque data.
ACPC Aligned Skull Template - [path]: Skull template to be used for ACPC alignment. It is not necessary to change this path unless you intend to use a non-standard template.
ACPC Aligned Brain Template - [path]: Brain template to be used for ACPC alignment. For human data, it can be ‘None’. It is not necessary to change this path unless you intend to use a non-standard template.
Configuration Without the GUI¶
The following nested key/value pairs will be set to these defaults if not defined in your pipeline configuration YAML.
anatomical_preproc:
acpc_alignment:
run: Off
# Run ACPC alignment before non-local means filtering or N4 bias
# correction
run_before_preproc: True
# ACPC size of brain in z-dimension in mm.
# Default: 150mm for human data.
brain_size: 150
# Choose a tool to crop the FOV in ACPC alignment.
# Using FSL's robustfov or flirt command.
# Default: robustfov for human data, flirt for monkey data.
FOV_crop: robustfov
# ACPC Target
# options: 'brain' or 'whole-head'
# note: 'brain' requires T1w_brain_ACPC_template below to be populated
acpc_target: 'whole-head'
# Run ACPC alignment on brain mask
# If the brain mask is in native space, turn it on
# If the brain mask is ACPC aligned, turn it off
align_brain_mask: Off
# ACPC aligned template
T1w_ACPC_template: /usr/share/fsl/5.0/data/standard/MNI152_T1_1mm.nii.gz
T1w_brain_ACPC_template: /usr/share/fsl/5.0/data/standard/MNI152_T1_1mm_brain.nii.gz
T2w_ACPC_template: None
T2w_brain_ACPC_template: None
Skull-Stripping¶
Skull-stripping is the removal of skull and other non-brain tissue like dura and eyes from anatomical images, which could otherwise complicate co-registration and normalization steps.
C-PAC provides options for configuring skull-stripping - users can select:
AFNI: 3dSkullStrip
FSL: BET. Further parameters for each of these tools are configurable.
niworkflows-ants: niworkflows’s antsBrainExtraction
U-Net: U-Net is a Fully Convolutional Network (FCN) for image segmentation. Users can now select this option for brain extraction, especially optimal for non-human primate data.
Providing their own brain mask for extraction
Configuring CPAC to run Skull-Stripping:¶
NOTE: If providing your own brain mask for extraction, you can leave the following options at default. The skull-stripping tools will not run if a brain mask is found in the data configuration file.
Already skull-stripped - [On,Off]: If inputs are already skull stripped (i.e. the structural input data is brain-only) then you can toggle this option to off.
Which tool for skull-stripping - [FSL, AFNI, niworkflows-ants]: Choose if you’d like to use FSL BET, AFNI 3dSkullStrip, or run all options in parallel.
Configuration Without the GUI¶
The following nested key/value pairs will be set to these defaults if not defined in your pipeline configuration YAML.
anatomical_preproc:
brain_extraction:
run: On
# using: ['3dSkullStrip', 'BET', 'UNet', 'niworkflows-ants', 'FreeSurfer-ABCD', 'FreeSurfer-BET-Tight', 'FreeSurfer-BET-Loose']
# this is a fork option
using: ['3dSkullStrip']
Configuring AFNI 3dSkullStrip options:¶
Note: These options are pre-set for AFNI 3dSkullStrip’s default values. These do not need to be modified unless you are looking to optimize the results of skull-stripping for your particular dataset.
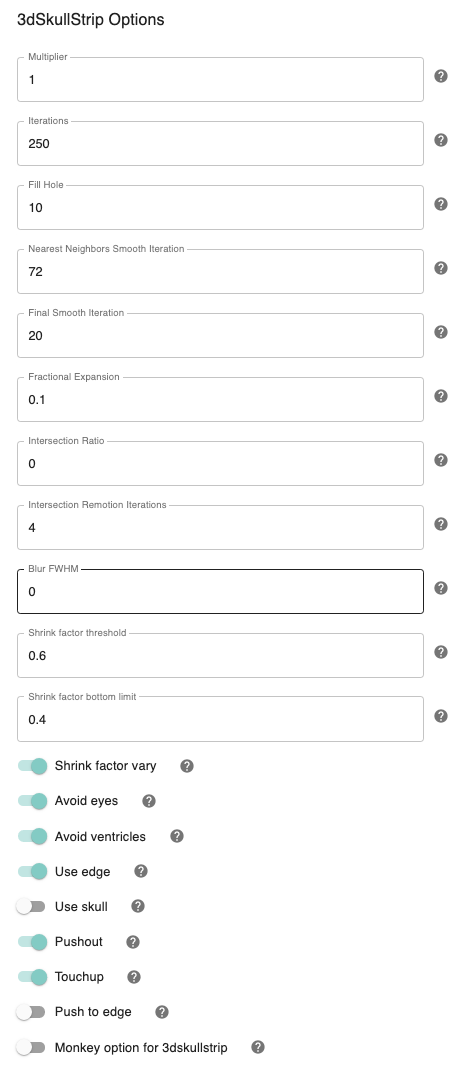
Shrink factor - [0.6]: Set the threshold value for controlling the brain vs non-brain voxels
Vary shrink factor - [On,Off]: Vary the shrink factor at every iteration of the algorithm. This prevents the likelihood of surface getting stuck in large pools of CSF before reaching the outer surface of the brain. Default is On.
Shrink factor bottom limit - [0.4]: The shrink factor bottom limit sets the lower threshold when varying the shrink factor. Default is 0.4, for when edge detection is used (which is On by default),otherwise the default value is 0.4.
Avoid ventricles - [On,Off]: Avoid ventricles while skull stripping. Default is On.
Number of iterations - [250]: Set the number of iterations. Default is 250. The number of iterations should depend upon the density of your mesh. Default is 250.
Pushout - [On, Off]: While expanding, consider the voxels above and not only the voxels below. Default is On.
Touchup - [On,Off]: Perform touchup operations at the end to include areas not covered by surface expansion.
Fill hole option - [10]: Give the maximum number of pixels on either side of the hole that can be filled. The default is 10 only if ’Touchup’ is On. Otherwise default is 0.
NN smooth - [72]: Perform final surface smoothing after all iterations. Default is 20.
Avoid eyes - [On,Off]: Avoid eyes while skull stripping. Default is On.
Use edge - [On,Off]: Use edge detection to reduce leakage into meninges. Default is On.
Fractional expansion - [0.1]: Speed of expansion. Default Value is 0.1
Push to edge - [Off,On]: Perform aggressive push to edge, this might cause leakage. Default is Off.
Use Skull - [Off, On]: Use the outer skull to limit expansion of surface into the skull in case of very strong shading artifacts. Use this only if you leakage into the skull.
Perc_init - [0]: Percentage of segments allowed to intersect surface.It is typically a number between 0 and 0.1, but can include negative values (which implies no testing for intersection). Default is 0.
Max_inter_iter - [4]: Number of iterations to remove intersection problems. With each iteration, the program automatically increases the amount of smoothing to get rid of intersections. Default is 4.
Fac - [1]: Multiply input dataset by FAC if range of values is too small. Default value is 1.
blur_fwhm - [2]: Blur datasets after spatial normalization. Default value is 2.
Configuration Without the GUI¶
The following nested key/value pairs will be set to these defaults if not defined in your pipeline configuration YAML.
anatomical_preproc:
brain_extraction:
AFNI-3dSkullStrip:
# Output a mask volume instead of a skull-stripped volume. The mask volume containes 0 to 6, which represents voxel's postion. If set to True, C-PAC will use this output to generate anatomical brain mask for further analysis.
mask_vol: False
# Set the threshold value controlling the brain vs non-brain voxels. Default is 0.6.
shrink_factor: 0.6
# Vary the shrink factor at every iteration of the algorithm. This prevents the likelihood of surface getting stuck in large pools of CSF before reaching the outer surface of the brain. Default is On.
var_shrink_fac: True
# The shrink factor bottom limit sets the lower threshold when varying the shrink factor. Default is 0.4, for when edge detection is used (which is On by default), otherwise the default value is 0.65.
shrink_factor_bot_lim: 0.4
# Avoids ventricles while skullstripping.
avoid_vent: True
# Set the number of iterations. Default is 250.The number of iterations should depend upon the density of your mesh.
n_iterations: 250
# While expanding, consider the voxels above and not only the voxels below
pushout: True
# Perform touchup operations at the end to include areas not covered by surface expansion.
touchup: True
# Give the maximum number of pixels on either side of the hole that can be filled. The default is 10 only if 'Touchup' is On - otherwise, the default is 0.
fill_hole: 10
# Perform nearest neighbor coordinate interpolation every few iterations. Default is 72.
NN_smooth: 72
# Perform final surface smoothing after all iterations. Default is 20.
smooth_final: 20
# Avoid eyes while skull stripping. Default is On.
avoid_eyes: True
# Use edge detection to reduce leakage into meninges and eyes. Default is On.
use_edge: True
# Speed of expansion.
exp_frac: 0.1
# Perform aggressive push to edge. This might cause leakage. Default is Off.
push_to_edge: False
# Use outer skull to limit expansion of surface into the skull in case of very strong shading artifacts. Use this only if you have leakage into the skull.
use_skull: Off
# Percentage of segments allowed to intersect surface. It is typically a number between 0 and 0.1, but can include negative values (which implies no testing for intersection).
perc_int: 0
# Number of iterations to remove intersection problems. With each iteration, the program automatically increases the amount of smoothing to get rid of intersections. Default is 4.
max_inter_iter: 4
# Multiply input dataset by FAC if range of values is too small.
fac: 1
# Blur dataset after spatial normalization. Recommended when you have lots of CSF in brain and when you have protruding gyri (finger like). If so, recommended value range is 2-4. Otherwise, leave at 0.
blur_fwhm: 0
# Set it as True if processing monkey data with AFNI
monkey: False
Configuring FSL BET options:¶
Note: These options are pre-set for FSL BET’s default values. These do not need to be modified unless you are looking to optimize the results of skull-stripping for your particular dataset.
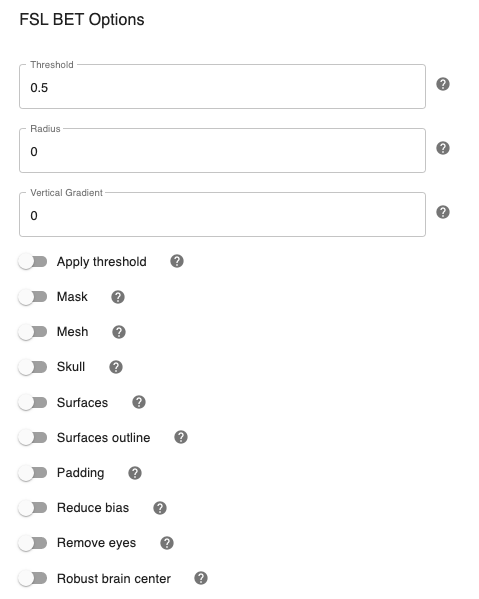
Threshold - [0.5]: Set the threshold value controlling the brain vs non-brain voxels. Default is 0.5
Radius - [0]: Integer value of head radius. Default is 0.
Vertical gradient - [Off,On]: Vertical gradient un fractional intensity threshold. Within the range of (-1,1).
Apply threshold - [Off,On]: Apply thresholding to segmented brain image and mask. Default is Off.
Mask - [Off, On]: Mask created along with skull stripping. Default option is On.
Mesh - [Off, On]: Mesh created along with skull stripping. Default is Off.
Skull - [Off,On]: Create a Skull Image. Default is Off.
Surfaces - [Off, On]: Get additional skull and scalp surfaces by running bet2 and betsurf. This is mutually exclusive with reduce bias, robust, padding, remove_eyes.
Surfaces outline - [Off, On]: Create a surface outline image, Default is Off.
Padding - [Off, On]: Add padding to the end of the image, improving BET. Mutually exclusive functional, reduce_bias, robust, padding, remove_eyes, surfaces.
Reduce bias - [Off, On]: Reduce bias and cleanup neck. Mutually exclusive with functional, reduce_bias, robust, padding, remove_eyes, surfaces.
Remove eyes - [Off,On]: Eyes and optic nerve cleanup. Mutually exclusive with functional, reduce_bias, robust, padding, remove_eyes, surfaces.
Robust brain center - [Off, On]: Robust brain center estimation. Mutually exclusive with functional, reduce_bias, robust, padding, remove_eyes, surfaces.
Configuration Without the GUI¶
The following nested key/value pairs will be set to these defaults if not defined in your pipeline configuration YAML.
anatomical_preproc:
brain_extraction:
FSL-BET:
# Set the threshold value controling the brain vs non-brain voxels, default is 0.5
frac: 0.5
# Mask created along with skull stripping. It should be `On`, if selected functionalMasking : ['Anatomical_Refined'] and `FSL` as skull-stripping method.
mask_boolean: On
# Mesh created along with skull stripping
mesh_boolean: Off
# Create a surface outline image
outline: Off
# Add padding to the end of the image, improving BET.Mutually exclusive with functional,reduce_bias,robust,padding,remove_eyes,surfaces
padding: Off
# Integer value of head radius
radius: 0
# Reduce bias and cleanup neck. Mutually exclusive with functional,reduce_bias,robust,padding,remove_eyes,surfaces
reduce_bias: Off
# Eyes and optic nerve cleanup. Mutually exclusive with functional,reduce_bias,robust,padding,remove_eyes,surfaces
remove_eyes: Off
# Robust brain center estimation. Mutually exclusive with functional,reduce_bias,robust,padding,remove_eyes,surfaces
robust: Off
# Create a skull image
skull: Off
# Gets additional skull and scalp surfaces by running bet2 and betsurf. This is mutually exclusive with reduce_bias, robust, padding, remove_eyes
surfaces: Off
# Apply thresholding to segmented brain image and mask
threshold: Off
# Vertical gradient in fractional intensity threshold (-1,1)
vertical_gradient : 0.0
Configuring niworkflows-ants options:¶
Note: These templates are used during niworkflows-ants skull stripping. e.g. OASIS template can be downloaded here.
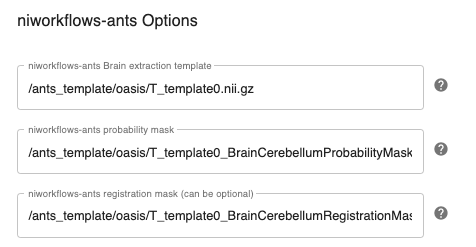
niworkflows_ants_template_path: Set the brain extraction template . e.g. OASIStemplate/T_template0_BrainCerebellumProbabilityMask.nii.gz
niworkflows_ants_mask_path: Set the brain extraction probability mask. e.g. OASIStemplate/T_template0_BrainCerebellumProbabilityMask.nii.gz
niworkflows_ants_regmask_path: Set the brain extraction registration mask, used for registration to limit the metric computation to a specific region. e.g. OASIStemplate/T_template0_BrainCerebellumRegistrationMask.nii.gz
Configuration Without the GUI¶
The following nested key/value pairs will be set to these defaults if not defined in your pipeline configuration YAML.
anatomical_preproc:
brain_extraction:
niworkflows-ants:
# Template to be used during niworkflows-ants.
# It is not necessary to change this path unless you intend to use a non-standard template.
# niworkflows-ants Brain extraction template
template_path : /ants_template/oasis/T_template0.nii.gz
# niworkflows-ants probability mask
mask_path : /ants_template/oasis/T_template0_BrainCerebellumProbabilityMask.nii.gz
# niworkflows-ants registration mask (can be optional)
regmask_path : /ants_template/oasis/T_template0_BrainCerebellumRegistrationMask.nii.gz
Anatomical Registration¶
In order to compare brain activations between subjects, individual functional and anatomical images must first be transformed to match a common template. The most commonly used template (MNI152) is maintained by the Montreal Neurological Institute, and is created by combining data from the brains of many different individuals to create an “average” brain. The image below shows how an individual brain is warped to match the shape of the template.
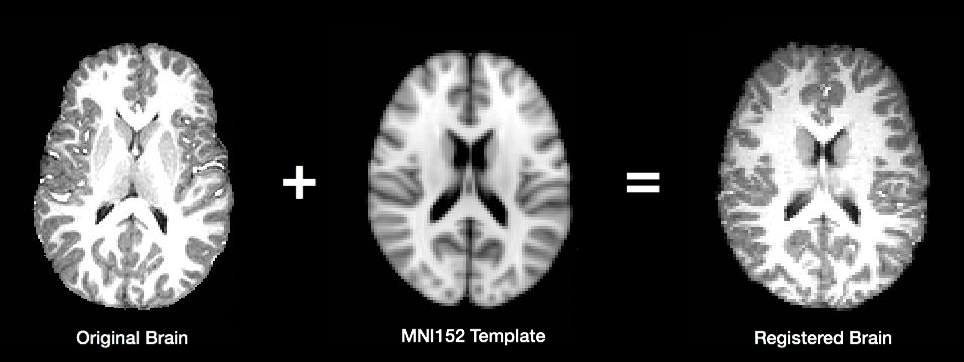
C-PAC provides the option of either using FSL (FLIRT and FNIRT) or Advanced Normalization Tools (ANTS) to register images. Although the use of ANTS requires an extra step during the C-PAC install process, we have found its results to be significantly better than those produced by FSL (a conclusion supported by a recent systematic analysis by Klein et al.).
During registration, individual anatomical images are first transformed to match the common template. Then, the functional data for each subject is registered to their own transformed anatomical image. Finally, functional derivative files are transformed to the common template. For more detail on how C-PAC computes these steps, please see the Registration Page of the developer documentation.
By default, C-PAC will register subject brains to the MNI152 template included with FSL. Users wishing to register their data to a different template (such as a group specific template) can specify alternative template files.
Configuring CPAC to Run Anatomical Registration¶
Anatomical Template Resolution - [1 An integer indicating three same dimensions (e.g., 1mm, 2mm, 3mm, 4mm); 2 A float number indicating three same dimensions (e.g., 3.5mm etc.); 3 Three numbers connected by ‘x’ indicating three different dimensions (e.g., 2.67mmx2.67mmx3mm etc.)]: The resolution to which anatomical images should be transformed during registration. This is the resolution at which processed anatomical files will be output.
Anatomical Template (Brain Only) - [path]: Template to be used during registration. It is not necessary to change this path unless you intend to use a non-standard template.
Anatomical Template (With Skull) - [path]: Template to be used during registration. It is not necessary to change this path unless you intend to use a non-standard template.
Anatomical to Template Registration Method - [ANTS, FSL, ANTS & FSL]: Registration method(s) to be used. Options are ANTS, FSL, or both.
ANTS skull-on transform - [Off, On]: Register skull-on anatomical image to template. Calculating the transform with skull-stripped images is reported to be better, but it requires very high-quality skull-stripping. If skull-stripping is imprecise, registration with skull is preferred. Note: This option only affects ANTS due to the fact that FNIRT already uses skull-on images for calculating warps.
Interpolation Method - [Linear, BSpline, LanczosWindowedSinc]: Interpolation method for writing out transformed anatomical images. ANTS registration tools only. Options are Linear, BSpline, or LanczosWindowedSinc.
ANTs Registration Parameters : Clicking on the setting icon will bring up a dialog where you can set ‘antsRegistration’ parameters.
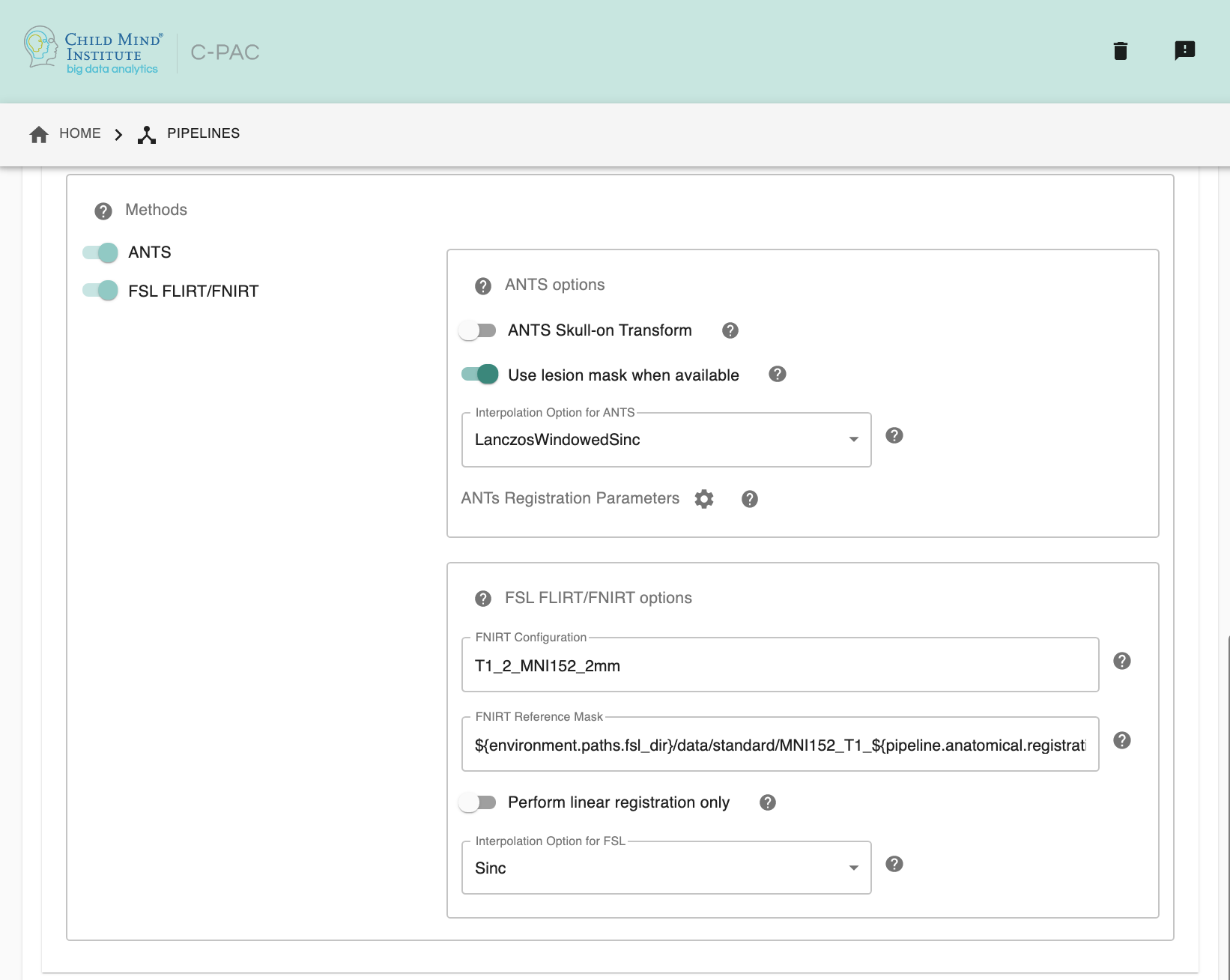
FNIRT Configuration - [path]: Configuration file specifying settings used during registration. Required if FSL is selected as the registration method. This file can be found in the
/etc/flirtschdirectory of your FSL install.FNIRT Reference Mask - [path]: A reference mask to be used by FNIRT.
Perform linear registration only - [Off, On]: Whether or not perform only FLIRT.
Interpolation Method - [trilinear, sinc, spline]: Interpolation method for writing out transformed anatomical images. FSL registration tools only. Options are trilinear, sinc, or spline.
Configuration Without the GUI¶
The following nested key/value pairs will be set to these defaults if not defined in your pipeline configuration YAML.
registration_workflows:
anatomical_registration:
run: On
# The resolution to which anatomical images should be transformed during registration.
# This is the resolution at which processed anatomical files will be output.
resolution_for_anat: 2mm
# Template to be used during registration.
# It is not necessary to change this path unless you intend to use a non-standard template.
T1w_brain_template: /usr/share/fsl/5.0/data/standard/MNI152_T1_${resolution_for_anat}_brain.nii.gz
# Template to be used during registration.
# It is not necessary to change this path unless you intend to use a non-standard template.
T1w_template: /usr/share/fsl/5.0/data/standard/MNI152_T1_${resolution_for_anat}.nii.gz
# Template to be used during registration.
# It is not necessary to change this path unless you intend to use a non-standard template.
T1w_brain_template_mask: /usr/share/fsl/5.0/data/standard/MNI152_T1_${resolution_for_anat}_brain_mask.nii.gz
# Register skull-on anatomical image to a template.
reg_with_skull: True
registration:
# using: ['ANTS', 'FSL', 'FSL-linear']
# this is a fork point
# selecting both ['ANTS', 'FSL'] will run both and fork the pipeline
using: ['ANTS']
# option parameters
ANTs:
# If a lesion mask is available for a T1w image, use it to improve the ANTs' registration
# ANTS registration only.
use_lesion_mask: False
# ANTs parameters for T1-template-based registration
T1_registration:
- collapse-output-transforms: 0
- dimensionality: 3
- initial-moving-transform :
initializationFeature: 0
- transforms:
- Rigid:
gradientStep : 0.1
metric :
type : MI
metricWeight: 1
numberOfBins : 32
samplingStrategy : Regular
samplingPercentage : 0.25
convergence:
iteration : 1000x500x250x100
convergenceThreshold : 1e-08
convergenceWindowSize : 10
smoothing-sigmas : 3.0x2.0x1.0x0.0
shrink-factors : 8x4x2x1
use-histogram-matching : True
- Affine:
gradientStep : 0.1
metric :
type : MI
metricWeight: 1
numberOfBins : 32
samplingStrategy : Regular
samplingPercentage : 0.25
convergence:
iteration : 1000x500x250x100
convergenceThreshold : 1e-08
convergenceWindowSize : 10
smoothing-sigmas : 3.0x2.0x1.0x0.0
shrink-factors : 8x4x2x1
use-histogram-matching : True
- SyN:
gradientStep : 0.1
updateFieldVarianceInVoxelSpace : 3.0
totalFieldVarianceInVoxelSpace : 0.0
metric:
type : CC
metricWeight: 1
radius : 4
convergence:
iteration : 100x100x70x20
convergenceThreshold : 1e-09
convergenceWindowSize : 15
smoothing-sigmas : 3.0x2.0x1.0x0.0
shrink-factors : 6x4x2x1
use-histogram-matching : True
winsorize-image-intensities :
lowerQuantile : 0.01
upperQuantile : 0.99
# Interpolation method for writing out transformed anatomical images.
# Possible values: Linear, BSpline, LanczosWindowedSinc
interpolation: LanczosWindowedSinc
FSL-FNIRT:
# Configuration file to be used by FSL to set FNIRT parameters.
# It is not necessary to change this path unless you intend to use custom FNIRT parameters or a non-standard template.
fnirt_config: T1_2_MNI152_2mm
# The resolution to which anatomical images should be transformed during registration.
# This is the resolution at which processed anatomical files will be output.
# specifically for monkey pipeline
ref_resolution: 2mm
# Reference mask for FSL registration.
ref_mask: /usr/share/fsl/5.0/data/standard/MNI152_T1_${resolution_for_anat}_brain_mask_dil.nii.gz
# Template to be used during registration.
# It is for monkey pipeline specifically.
FNIRT_T1w_brain_template: None
# Template to be used during registration.
# It is for monkey pipeline specifically.
FNIRT_T1w_template: None
# Interpolation method for writing out transformed anatomical images.
# Possible values: trilinear, sinc, spline
interpolation: sinc
# Identity matrix used during FSL-based resampling of anatomical-space data throughout the pipeline.
# It is not necessary to change this path unless you intend to use a different template.
identity_matrix: /usr/share/fsl/5.0/etc/flirtsch/ident.mat
# Reference mask for FSL registration.
ref_mask: /usr/share/fsl/5.0/data/standard/MNI152_T1_${resolution_for_anat}_brain_mask_dil.nii.gz
# Reference mask with 2mm resolution to be used during FNIRT-based brain extraction in ABCD-options pipeline.
ref_mask_res-2: /usr/share/fsl/5.0/data/standard/MNI152_T1_2mm_brain_mask_dil.nii.gz
# Template with 2mm resolution to be used during FNIRT-based brain extraction in ABCD-options pipeline.
T1w_template_res-2: /usr/share/fsl/5.0/data/standard/MNI152_T1_2mm.nii.gz
overwrite_transform:
run: Off
# Choose the tool to overwrite transform, currently only support 'FSL' to overwrite 'ANTs' transforms in ABCD-options pipeline.
# using: 'FSL'
using: FSL
Anatomical Tissue Segmentation¶
C-PAC uses FSL/FAST to automatically segment brain images into white matter, gray matter, and CSF. This is done using probability maps that contain information about the likelihood that a given voxel will be of a particular tissue type. Users specify a probability threshold such that voxels meeting a minimum probability of being a particular tissue will be classified as such. This results in masks containing voxels of only a single tissue type.
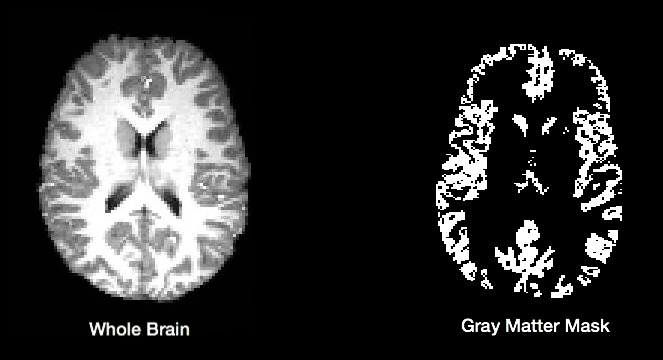
The default tissue probability maps (referred to as Prior Probability Maps) used during segmentation are based on information from a large number of brains, and are based on the priors distributed with FSL and are included in the “Image Resource Files” package downloaded during installation. Also, CPAC has thresholding and erosion options for anatomical segmentation to further refine the resulting segmentation tissue masks. Threshold value and erosion proportion can be changeable by user. The erosion implementation is adapted from fmriprep.
For more detail on how CPAC computes these steps, please see the Segmentation Page of the developer documentation.
Thresholding options have returned, and new erosion options for anatomical segmentation have been introduced. The erosion implementation was adapted from fmriprep.
If you would like to use different priors, they must first be binarized such that for each voxel the probability for each tissue type is set to either 0% or 100%.
The following bash script will binarize existing priors:
# Define what kind of priors to generate (gray, white, or csf)
tissue=gray
# Define threshold to use when binarizing data
threshold=0.5
# Copy existing priors (in this example, from FSL)
3dcopy $FSL_DIR/data/standard/tissuepriors/avg152T1_${tissue}.hdr avg152T1_${tissue}.nii.gz
# Binarize image using threshold set above
fslmaths avg152T1_${tissue}.nii.gz -thr $threshold -bin avg152T1_${tissue}_2mm_bin
In addition, C-PAC offers template-based segmentation options that facilitate nonhuman data processing. Optimal for use with functional-only pipelines commonly used for rodent data, users can now employ a template-based tissue segmentation approach that applies inverse registration transforms to template-space tissue priors.
C-PAC offers ANTs prior-based tissue segmentation, which is optimal for non-human primate segmentations. Users could provide atlas and atlas segmentation images to perform ANTs Prior-based Segmentation.
Configuring CPAC to Run Anatomical Tissue Segmentation¶
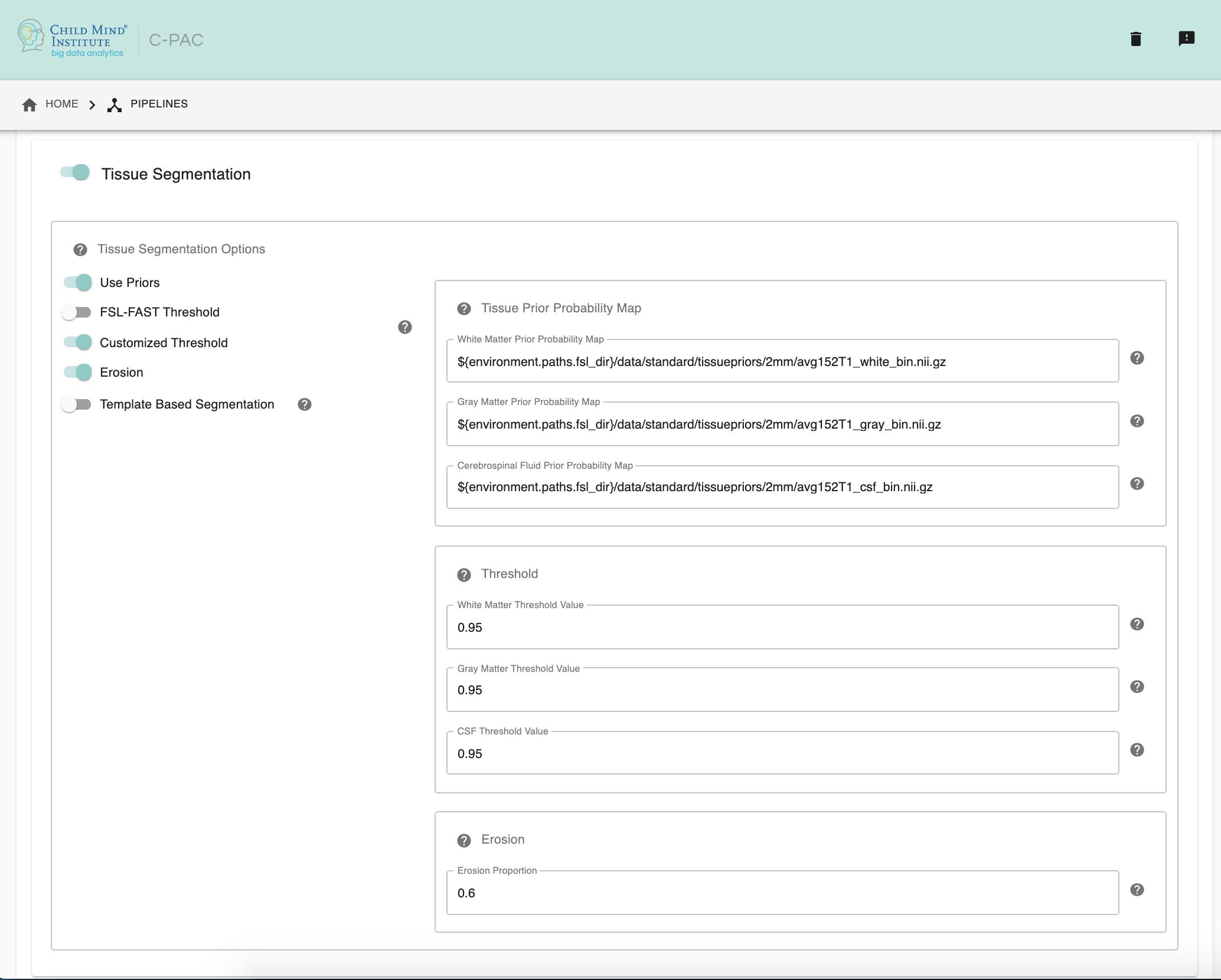
Tissue Segmentation - [On, Off]: Automatically segment anatomical images into white matter, gray matter, and CSF based on prior probability maps.
Use Priors - [On, Off]: Whether or not to use template-space tissue priors to refine the binary tissue masks generated by segmentation.
White Matter Prior Probability Map - [path]: Full path to a binarized White Matter prior probability map. It is not necessary to change this path unless you intend to use non-standard priors.
Gray Matter Prior Probability Map - [path]: Full path to a binarized Gray Matter prior probability map. It is not necessary to change this path unless you intend to use non-standard priors.
CSF Prior Probability Map - [path]: Full path to a binarized CSF prior probability map. It is not necessary to change this path unless you intend to use non-standard priors.
FSL-FAST Thresholding - [On, Off]]: Use FSL-FAST generated binary masks to generate the resulting segmentation tissue masks.
Customized Thresholding - [On,Off]]: Set the threshold value for tissue probability maps to generate the resulting segmentation tissue masks.
White Matter Threshold Value - [float]: Set the threshold value for refining the resulting White Matter segmentation tissue mask, if choose Customized Thresholding. The default value is 0.95.
Gray Matter Threshold Value - [float]: Set the threshold value for refining the resulting Gray Matter segmentation tissue mask, if choose Customized Thresholding. The default value is 0.95.
CSF Threshold Value - [float]: Set the threshold value for refining the resulting CSF segmentation tissue mask, if choose Customized Thresholding. The default value is 0.95.
Erosion - [On, Off]: Whether or not to use erosion to erode binarized tissue masks.
Erosion Proportion - [float]: Set the erosion proportion, if use erosion to erode binarized tissue masks. The default is 0.6.
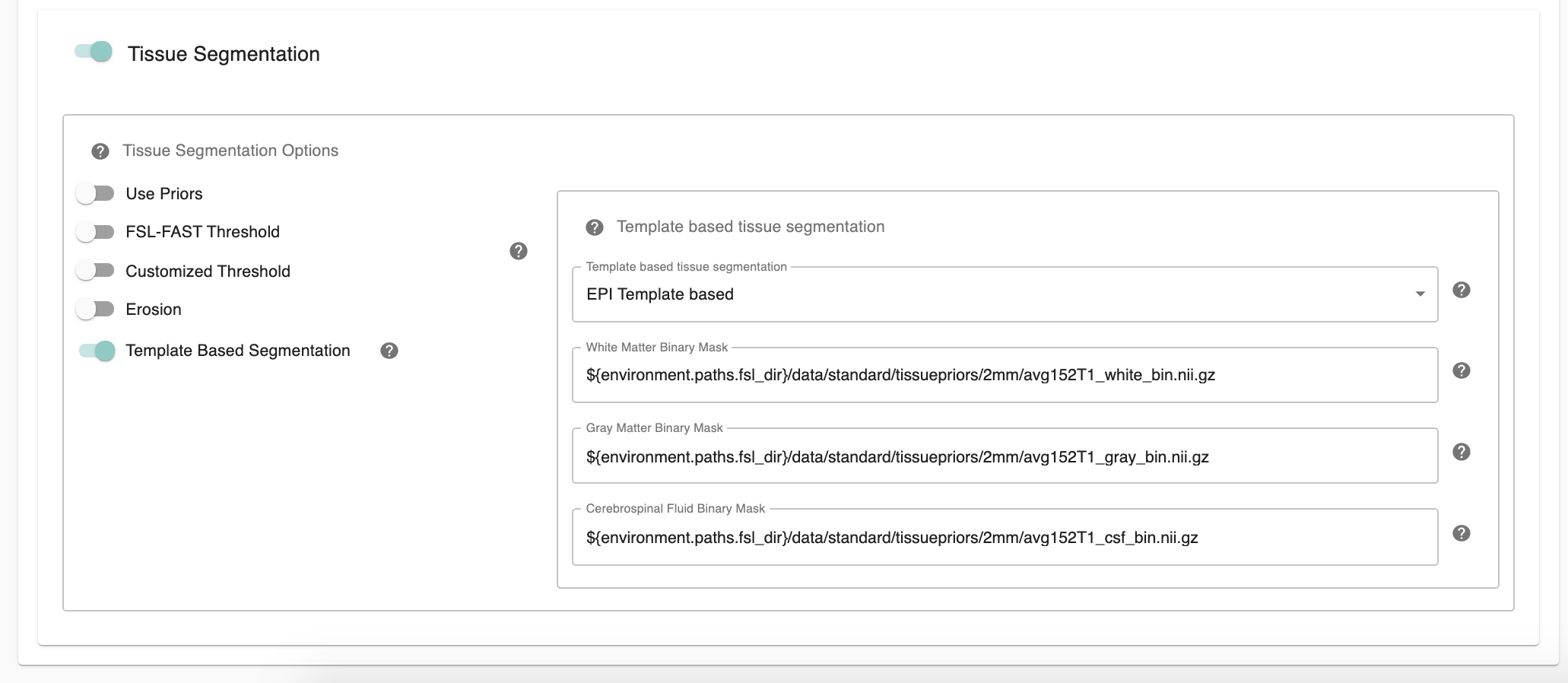
Template Based Segmentation - [EPI Template based, T1 Template based]: Optimal for use with functional-only pipelines commonly used for rodent data, users can now employ a template-based tissue segmentation approach that applies inverse registration transforms to template-space tissue priors. If choose ‘EPI Template based’ or ‘T1 Template based’ as template based segmentation method, please make sure to specify white matter, gray matter, CSF mask paths at below three configurations.
White Matter Binary Mask - [path]: Full path to a binarized White Matter mask.
Gray Matter Binary Mask - [path]: Full path to a binarized Gray Matter mask.
CSF Prior Binary Mask - [path]: Full path to a binarized CSF mask.
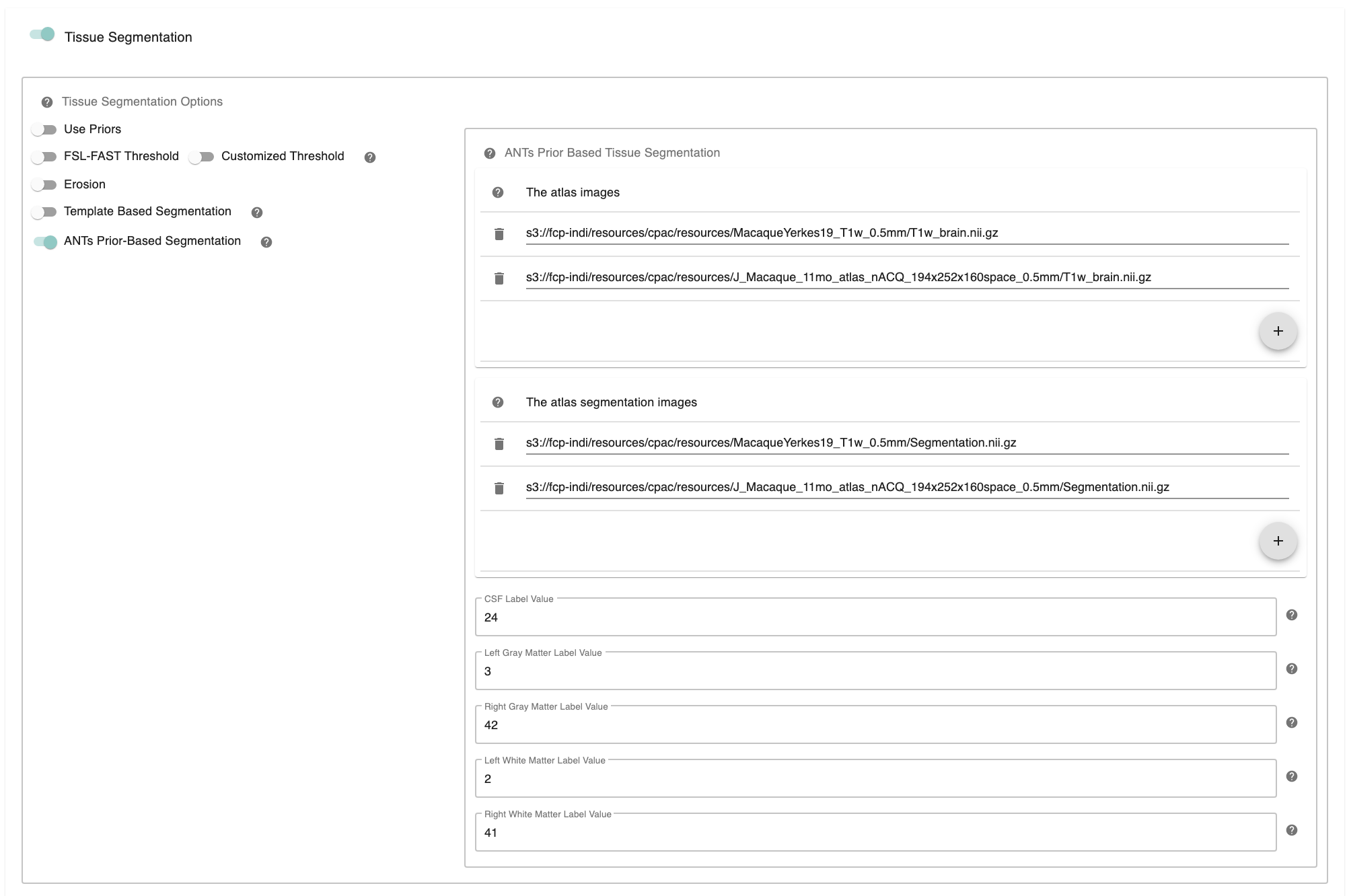
ANTs Prior-Based Segmentation - [On, Off]: ANTs Prior-based Segmentation workflow that has shown optimal results for non-human primate data. Generate white matter, gray matter, CSF masks based on antsJointLabelFusion.
The atlas image - [path]: The atlas image assumed to be used in ANTs Prior-based Segmentation. Clicking on the + icon to the right of the box here will bring up a dialog where you can define multiple paths to NifTIs containing the atlas image. You may add multiple images to the box.
The atlas segmentation images - [path]: The number of specified segmentations should be identical to the number of atlas brain images. Clicking on the + icon to the right of the box here will bring up a dialog where you can define multiple paths to NifTIs containing the atlas segmentation image. You may add multiple images to the box.
CSF Label Value - [integer]: Label value corresponding to CSF in multiatlas file. It is not necessary to change this values unless your CSF/GM/WM label values are different from Freesurfer Color Lookup Table.
Left Gray Matter Label Value - [integer]: Label value corresponding to Left Gray Matter in multiatlas file. It is not necessary to change this values unless your CSF/GM/WM label values are different from Freesurfer Color Lookup Table.
Right Gray Matter Label Value - [integer]: Label value corresponding to Right Gray Matter in multiatlas file. It is not necessary to change this values unless your CSF/GM/WM label values are different from Freesurfer Color Lookup Table.
Left White Matter Label Value - [integer]: Label value corresponding to Left White Matter in multiatlas file. It is not necessary to change this values unless your CSF/GM/WM label values are different from Freesurfer Color Lookup Table.
Right White Matter Label Value - [integer]: Label value corresponding to Right White Matter in multiatlas file. It is not necessary to change this values unless your CSF/GM/WM label values are different from Freesurfer Color Lookup Table.
Configuration Without the GUI¶
The following nested key/value pairs will be set to these defaults if not defined in your pipeline configuration YAML.
segmentation:
# Automatically segment anatomical images into white matter, gray matter,
# and CSF based on prior probability maps.
run: On
tissue_segmentation:
# using: ['FSL-FAST', 'Template_Based', 'ANTs_Prior_Based', 'FreeSurfer']
# this is a fork point
using: ['FSL-FAST']
# option parameters
FSL-FAST:
thresholding:
# thresholding of the tissue segmentation probability maps
# options: 'Auto', 'Custom'
use: 'Auto'
Custom:
# Set the threshold value for the segmentation probability masks (CSF, White Matter, and Gray Matter)
# The values remaining will become the binary tissue masks.
# A good starting point is 0.95.
# CSF (cerebrospinal fluid) threshold.
CSF_threshold_value : 0.95
# White matter threshold.
WM_threshold_value : 0.95
# Gray matter threshold.
GM_threshold_value : 0.95
use_priors:
# Use template-space tissue priors to refine the binary tissue masks generated by segmentation.
run: On
# Full path to a directory containing binarized prior probability maps.
# These maps are included as part of the 'Image Resource Files' package available on the Install page of the User Guide.
# It is not necessary to change this path unless you intend to use non-standard priors.
priors_path: $FSLDIR/data/standard/tissuepriors/2mm
# Full path to a binarized White Matter prior probability map.
# It is not necessary to change this path unless you intend to use non-standard priors.
WM_path: $priors_path/avg152T1_white_bin.nii.gz
# Full path to a binarized Gray Matter prior probability map.
# It is not necessary to change this path unless you intend to use non-standard priors.
GM_path: $priors_path/avg152T1_gray_bin.nii.gz
# Full path to a binarized CSF prior probability map.
# It is not necessary to change this path unless you intend to use non-standard priors.
CSF_path: $priors_path/avg152T1_csf_bin.nii.gz
Template_Based:
# These masks should be in the same space of your registration template, e.g. if
# you choose 'EPI Template' , below tissue masks should also be EPI template tissue masks.
#
# Options: ['T1_Template', 'EPI_Template']
template_for_segmentation: ['T1_Template']
# These masks are included as part of the 'Image Resource Files' package available
# on the Install page of the User Guide.
# Full path to a binarized White Matter mask.
WHITE: $FSLDIR/data/standard/tissuepriors/2mm/avg152T1_white_bin.nii.gz
# Full path to a binarized Gray Matter mask.
GRAY: $FSLDIR/data/standard/tissuepriors/2mm/avg152T1_gray_bin.nii.gz
# Full path to a binarized CSF mask.
CSF: $FSLDIR/data/standard/tissuepriors/2mm/avg152T1_csf_bin.nii.gz
ANTs_Prior_Based:
# Generate white matter, gray matter, CSF masks based on antsJointLabelFusion
# ANTs Prior-based Segmentation workflow that has shown optimal results for non-human primate data.
# The atlas image assumed to be used in ANTs Prior-based Segmentation.
template_brain_list :
- /cpac_templates/MacaqueYerkes19_T1w_0.5mm_desc-JLC_T1w_brain.nii.gz
- /cpac_templates/J_Macaque_11mo_atlas_nACQ_194x252x160space_0.5mm_desc-JLC_T1w_brain.nii.gz
# The atlas segmentation images.
# For performing ANTs Prior-based segmentation method
# the number of specified segmentations should be identical to the number of atlas brain image sets.
# eg.
# ANTs_prior_seg_template_brain_list :
# - atlas1.nii.gz
# - atlas2.nii.gz
# ANTs_prior_seg_template_segmentation_list:
# - segmentation1.nii.gz
# - segmentation1.nii.gz
template_segmentation_list:
- /cpac_templates/MacaqueYerkes19_T1w_0.5mm_desc-JLC_Segmentation.nii.gz
- /cpac_templates/J_Macaque_11mo_atlas_nACQ_194x252x160space_0.5mm_desc-JLC_Segmentation.nii.gz
# Label values corresponding to CSF/GM/WM in atlas file
# It is not necessary to change this values unless your CSF/GM/WM label values are different from Freesurfer Color Lookup Table.
# https://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/AnatomicalROI/FreeSurferColorLUT
# Label values corresponding to CSF in multiatlas file
CSF_label : [24]
# Label values corresponding to Gray Matter in multiatlas file
GM_label : [3, 42]
# Label values corresponding to White Matter in multiatlas file
WM_label : [2, 41]
FreeSurfer:
# Use mri_binarize --erode option to erode segmentation masks
erode: 0
# Label values corresponding to CSF in FreeSurfer aseg segmentation file
CSF_label : [24]
# Label values corresponding to Gray Matter in FreeSurfer aseg segmentation file
GM_label : [3, 42]
# Label values corresponding to White Matter in FreeSurfer aseg segmentation file
WM_label : [2, 41]
References¶
Smith, Stephen M., Fast robust automated brain extraction, Human Brain Mapping 2002, Volume 17 Issue 3, page 143-155.
Tustison et al., N4ITK: Improved N3 Bias Correction, IEEE Transactions on Medical Imaging, 29(6):1310-1320, June 2010.
