Timeseries Extraction¶
C-PAC lets you easily export BOLD timeseries in a number of different ways. This can be useful for those wishing to undertake advanced analysis not included in C-PAC, but still take advantage of its robust pre-processing features. For instructions on how to use these seeds within C-PAC, please see Seed-based Correlation Analysis.
ROI Timeseries Extraction allows you to export the timeseries for one or more regions of interest (ROIs). This is done by calculating the average timeseries across all voxels within an ROI. As such, C-PAC will output one timeseries for each ROI specified by you.
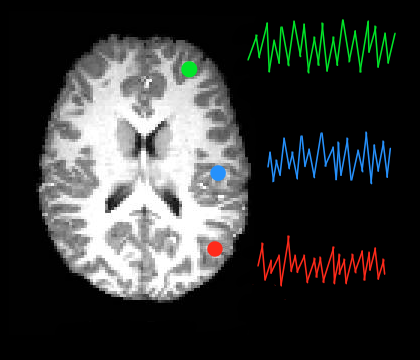
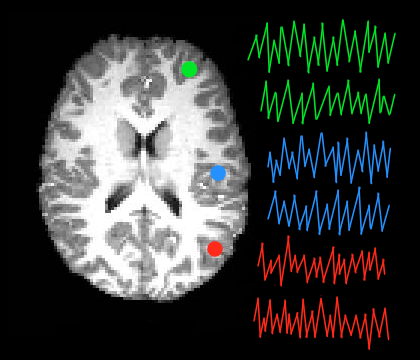
When an ROI is placed within a functionally homogeneous area, averaging signals in this way can produce a timeseries which may more accurately reflect the overall activity pattern in the region than does the timeseries of any individual voxel. Voxel Timeseries Extraction will export the individual timeseries of all voxels within one or more masks.
Configuring ROI Time Series Extraction¶
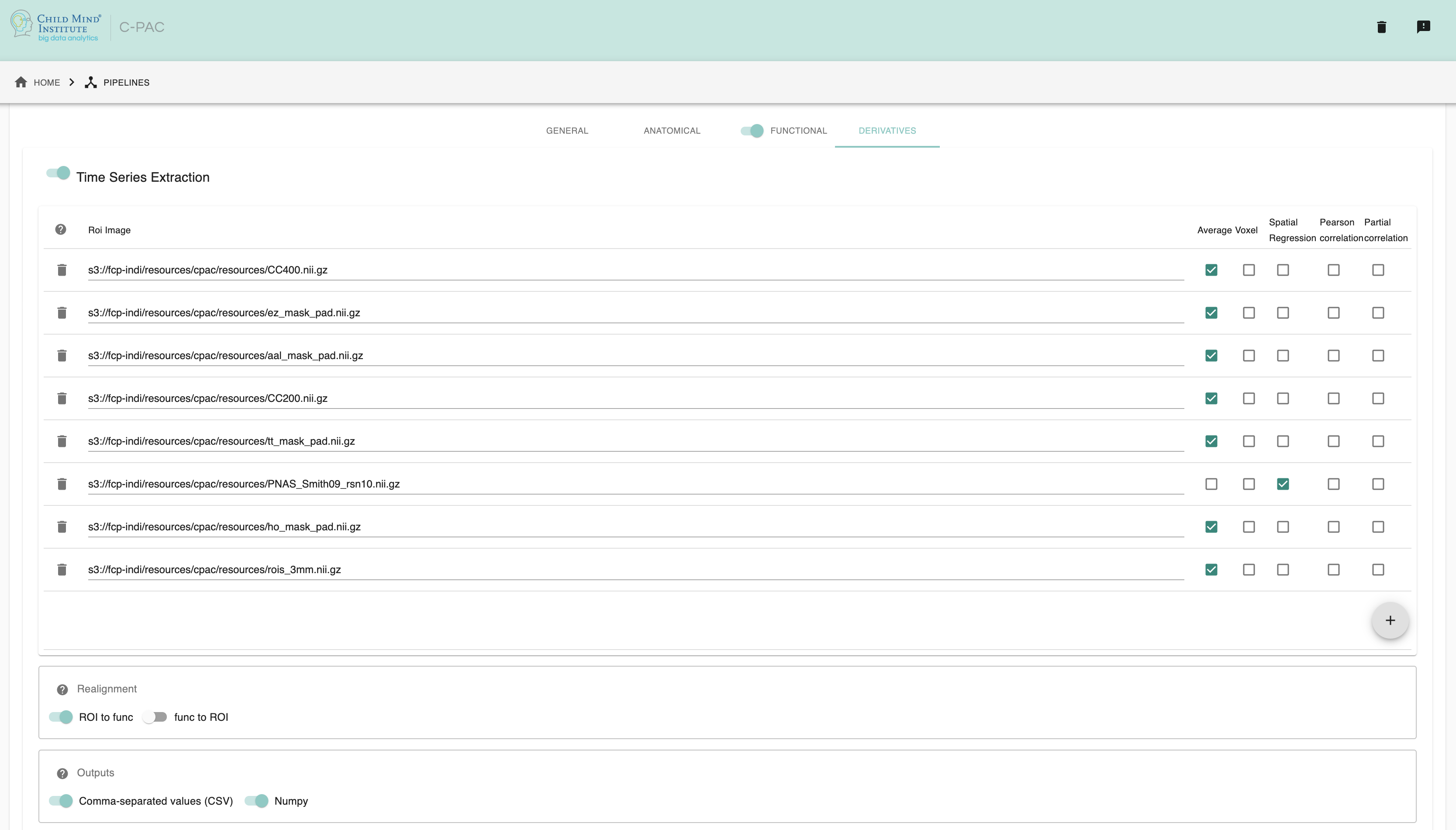
Time Series Extraction- [On,Off]: Extract the average time series of one or more ROIs/seeds. Must be enabled if you wish to run seed-based correlation analysis.
- TSE ROI Image - [path]: Clicking on the + icon to the bottom-right of the box here will bring up a dialog where you can define multiple paths to NifTIs containing ROI masks. You may add multiple ROIs to the box. Three columns within the box can be checked on and off to enable specific types of TSE:
Avg - For each ROI, output the average of the all the voxel time series within that ROI.
Voxel - For each ROI, output the individual voxel time series for all voxels within that ROI.
SpatialReg - Use a spatial map as a spatial regressor in a GLM to find the time series associated with the voxels in that map (see dual regression).
Realignment - [ROI to func, func to ROI]: Choose functional time-series and ROI realignment method. ‘ROI to func’ will realign the atlas/ROI to functional space (fast). ‘func to ROI’ will realign the functional time series to the atlas/ROI space. NOTE: in rare cases, realigning the ROI to the functional space may result in small misalignments for very small ROIs - please double check your data if you see issues.
Connectivity Matrix: A connectivity matrix can be generated via nilearn [1, 2, 3], ndmg [4, 5] or AFNI for the
Avgtimeseries output.
Configuration Without the GUI¶
The following nested key/value pairs will be set to these defaults if not defined in your pipeline configuration YAML.
Unlike most lines in a C-PAC pipeline configuration file, the keys under *_roi_paths are user-definable.
If you exclude *_roi_paths from your pipeline configuration, the default atlases and analyses will be used.
If you include *_roi_paths, only the atlases and analyses provided will be used, unless you add the optional key/value pair roi_paths_fully_specified: False in the relevant section, in which case the default atlases and analyses will be updated with the atlases and analyses specified in your pipeline config. For example,
timeseries_extraction:
roi_paths_fully_specified: False
seed_based_correlation_analysis:
roi_paths_fully_specified: False
would keep any default atlases not respecified in your pipeline configuration.
timeseries_extraction:
run: On
# Enter paths to region-of-interest (ROI) NIFTI files (.nii or .nii.gz) to be used for time-series extraction, and then select which types of analyses to run.
# Denote which analyses to run for each ROI path by listing the names below. For example, if you wish to run Avg and SpatialReg, you would enter: '/path/to/ROI.nii.gz': Avg, SpatialReg
# available analyses:
# /path/to/atlas.nii.gz: Avg, Voxel, SpatialReg
tse_roi_paths:
/cpac_templates/CC400.nii.gz: Avg
/cpac_templates/aal_mask_pad.nii.gz: Avg
/cpac_templates/CC200.nii.gz: Avg
/cpac_templates/tt_mask_pad.nii.gz: Avg
/cpac_templates/PNAS_Smith09_rsn10.nii.gz: SpatialReg
/cpac_templates/ho_mask_pad.nii.gz: Avg
/cpac_templates/rois_3mm.nii.gz: Avg
/ndmg_atlases/label/Human/AAL_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/CAPRSC_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/DKT_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/DesikanKlein_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/HarvardOxfordcort-maxprob-thr25_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/HarvardOxfordsub-maxprob-thr25_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/Juelich_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/MICCAI_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/cpac_templates/Schaefer2018_space-FSLMNI152_res-2mm_desc-1000Parcels17NetworksOrder.nii.gz: Avg
/cpac_templates/Schaefer2018_space-FSLMNI152_res-2mm_desc-200Parcels17NetworksOrder.nii.gz: Avg
/cpac_templates/Schaefer2018_space-FSLMNI152_res-2mm_desc-300Parcels17NetworksOrder.nii.gz: Avg
/cpac_templates/Schaefer2018_space-FSLMNI152_res-2mm_desc-400Parcels17NetworksOrder.nii.gz: Avg
/ndmg_atlases/label/Human/Talairach_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/Brodmann_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/Desikan_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/Glasser_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/Slab907_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/Yeo-17-liberal_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/Yeo-17_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/Yeo-7-liberal_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
/ndmg_atlases/label/Human/Yeo-7_space-MNI152NLin6_res-1x1x1.nii.gz: Avg
# Functional time-series and ROI realignment method: ['ROI_to_func'] or ['func_to_ROI']
# 'ROI_to_func' will realign the atlas/ROI to functional space (fast)
# 'func_to_ROI' will realign the functional time series to the atlas/ROI space
#
# NOTE: in rare cases, realigning the ROI to the functional space may
# result in small misalignments for very small ROIs - please double
# check your data if you see issues
realignment: 'ROI_to_func'
connectivity_matrix:
# Create a connectivity matrix from timeseries data
# Options:
# ['AFNI', 'Nilearn', 'ndmg']
using:
- Nilearn
- ndmg
# Options:
# ['Pearson', 'Partial']
# Note: These options are not configurable for ndmg, which will ignore these options
measure:
- Pearson
- Partial
References
Abraham, A., Pedregosa, F., Eickenberg, M., Gervais, P., Mueller, A., Kossaifi, J., Gramfort, A., Thirion, B., and Varoquaux, G. 2014. Machine learning for neuroimaging with scikit-learn. Frontiers in neuroinformatics, 8 , pp. 14 doi:10.3389/fninf.2014.00014
The nilearn developers. 2021. 3.1.1.2. Extracting signals on a parcellation. Nilearn: Statistics for neuroimaging in Python: User guide
The nilearn developers. 2021. 8.1.1. Nilearn.Connectome.ConnectivityMeasure. Nilearn: Statistics for neuroimaging in Python: User guide
Kiar, G., Bridgeford, E. W., Roncal, W. R. G., Consortium for Reliability and Reproducibility (CoRR), Chandrashekhar, V., Mhembere, D., Ryman, S., Zuo, X.-N., Margulies, D. S., Craddock, R. C., Priebe, C. E., Jung, R., Calhoun, V. D., Caffo, B., Burns, R., Milham, M. P., and Vogelstein, J. T. 2018 April. A High-Throughput Pipeline Identifies Robust Connectomes But Troublesome Variability. doi:10.1101/188706
NeuroData. 2018 January. ndmg v0.1.0.