1. Tutorial: optimizing memory estimation¶
(You can ⇩ download this notebook 💾 and run it locally.)
C-PAC has some built-in heuristics for estimating the memory required for each node, but these estimates can be greatly improved by providing observed memory usage for a given configuration and comparable data. This tutorial will step through the process of an observation run and an optimized run.
Note: The code cells in this notebook are using a BASH kernel and cpac (Python package) v0.5.0
1.1. Observation run¶
First, we need to run a single exemplar through our pipeline configuration to observe how much memory is used at each node. How many cores we provide to this run should be the same number we intend to use in our optimized runs, but we’ll want to give this run lots of memory to allow for the built-in conservative memory estimates.
For this tutorial, we’ll use a subject from ADHD-200, from the FCP-INDI AWS S3 bucket, and we’ll use the anatomical only preconfiguration.
[1]:
cpac run \
s3://fcp-indi/data/Projects/ADHD200/RawDataBIDS/NYU cpac_runs/observation participant \
--preconfig anat-only \
--participant_label 0010042 \
--n_cpus 4 \
--mem_gb 10
Loading 🐳 Docker
[…]
220414-15:17:19,750 nipype.workflow INFO:
Running in parallel.
220414-15:17:19,751 nipype.workflow WARNING:
The following nodes are estimated to exceed the total amount of memory available (10.00GB):
resample_u: 14.799999999999999 GB
resample_o: 14.799999999999999 GB
resample_u: 14.799999999999999 GB
resample_o: 14.799999999999999 GB
220414-15:17:20,8 nipype.workflow INFO:
Error of subject workflow cpac_sub-0010042_ses-1
CPAC run error:
Pipeline configuration: cpac_anat
Subject workflow: cpac_sub-0010042_ses-1
[…]
RuntimeError: Insufficient resources available for job:
resample_u: 14.799999999999999 GB
resample_o: 14.799999999999999 GB
resample_u: 14.799999999999999 GB
resample_o: 14.799999999999999 GB
[…]
We can see the initial estimate requires at least 14.8 GB for some nodes, so we’ll need to give it more memory. We have a couple options here.
If we don’t have hard memory limits (we expect our system will allow us to use more memory than we allocate), we can tell C-PAC to ignore the insufficient resources by adjusting our pipeline configuration, like
FROM: anat-only
pipeline_setup:
system_config:
raise_insufficient: Off
[2]:
cpac run \
s3://fcp-indi/data/Projects/ADHD200/RawDataBIDS/NYU cpac_runs/insufficient participant \
--pipeline_file $PWD/configs/pipeline/anat-only-insufficient.yml \
--participant_label 0010042 \
--n_cpus 4 \
--mem_gb 10
Loading 🐳 Docker
[…]
Skipping bids-validator for S3 datasets...
Loading the 'anat-only' pre-configured pipeline.
#### Running C-PAC for 0010042
Number of participants to run in parallel: 1
Input directory: s3://fcp-indi/data/Projects/ADHD200/RawDataBIDS/NYU
Output directory: […]/cpac_runs/insufficient/output
Working directory: […]/cpac_runs/insufficient/working
Log directory: […]/cpac_runs/insufficient/log
Remove working directory: False
Available memory: 10.0 (GB)
Available threads: 4
Number of threads for ANTs: 1
Parsing s3://fcp-indi/data/Projects/ADHD200/RawDataBIDS/NYU..
Connecting to AWS: fcp-indi anonymously...
gathering files from S3 bucket (s3.Bucket(name='fcp-indi')) for data/Projects/ADHD200/RawDataBIDS/NYU
Starting participant level processing
Run called with config file […]/cpac_runs/insufficient/cpac_pipeline_config_2022-04-14T15-18-12Z.yml
220414-15:18:19,245 nipype.workflow INFO:
Run command: run s3://fcp-indi/data/Projects/ADHD200/RawDataBIDS/NYU cpac_runs/insufficient participant --pipeline_file […]/configs/pipeline/anat-only-insufficient.yml --participant_label 0010042 --n_cpus 4 --mem_gb 10
C-PAC version: 1.8.4.dev
Setting maximum number of cores per participant to 4
Setting number of participants at once to 1
Setting OMP_NUM_THREADS to 1
Setting MKL_NUM_THREADS to 1
Setting ANTS/ITK thread usage to 1
Maximum potential number of cores that might be used during this run: 4
[…]
End of subject workflow cpac_sub-0010042_ses-1
CPAC run complete:
Pipeline configuration: cpac_anat
Subject workflow: cpac_sub-0010042_ses-1
[…]
Or
We can just allocate enough memory to satisfy the estimates
[3]:
cpac run \
s3://fcp-indi/data/Projects/ADHD200/RawDataBIDS/NYU cpac_runs/enough participant \
--preconfig anat-only \
--participant_label 0010042 \
--n_cpus 4 \
--mem_gb 14.8
Loading 🐳 Docker
[…]
Skipping bids-validator for S3 datasets...
Loading the 'anat-only' pre-configured pipeline.
#### Running C-PAC for 0010042
Number of participants to run in parallel: 1
Input directory: s3://fcp-indi/data/Projects/ADHD200/RawDataBIDS/NYU
Output directory: […]/cpac_runs/enough/output
Working directory: […]/cpac_runs/enough/working
Log directory: […]/cpac_runs/enough/log
Remove working directory: False
Available memory: 14.8 (GB)
Available threads: 4
Number of threads for ANTs: 1
Parsing s3://fcp-indi/data/Projects/ADHD200/RawDataBIDS/NYU..
Connecting to AWS: fcp-indi anonymously...
gathering files from S3 bucket (s3.Bucket(name='fcp-indi')) for data/Projects/ADHD200/RawDataBIDS/NYU
Starting participant level processing
Run called with config file […]/cpac_runs/enough/cpac_pipeline_config_2022-04-14T15-25-19Z.yml
220414-15:25:26,762 nipype.workflow INFO:
Run command: run s3://fcp-indi/data/Projects/ADHD200/RawDataBIDS/NYU cpac_runs/enough participant --preconfig anat-only --participant_label 0010042 --n_cpus 4 --mem_gb 14.8
C-PAC version: 1.8.4.dev
Setting maximum number of cores per participant to 4
Setting number of participants at once to 1
Setting OMP_NUM_THREADS to 1
Setting MKL_NUM_THREADS to 1
Setting ANTS/ITK thread usage to 1
Maximum potential number of cores that might be used during this run: 4
[…]
End of subject workflow cpac_sub-0010042_ses-1
CPAC run complete:
Pipeline configuration: cpac_anat
Subject workflow: cpac_sub-0010042_ses-1
[…]
Now we have a callback.log file with observed memory usage. We can peek at the observations with cpac parse-resources.
[4]:
cpac parse-resources --help
usage: cpac parse-resources [-h]
[--filter_field {runtime,estimate,efficiency}]
[--filter_group {lowest,highest}]
[--filter_count FILTER_COUNT]
callback
positional arguments:
callback callback.log file found in the 'log' directory of the
specified derivatives path
optional arguments:
-h, --help show this help message and exit
--filter_field {runtime,estimate,efficiency}, -f {runtime,estimate,efficiency}
--filter_group {lowest,highest}, -g {lowest,highest}
--filter_count FILTER_COUNT, -n FILTER_COUNT
Looking at the 5 most efficient nodes in each successful run, we can see the resource usage varies from run to run, even with the same data, system, and configuration:
[5]:
cpac parse-resources \
--filter_field efficiency \
--filter_group highest \
--filter_count 5 \
cpac_runs/insufficient/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┓
┃ ┃ Memory ┃ Memory ┃ Memory ┃
┃ Task ID ┃ Used ┃ Estimated ┃ Efficien… ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━┩
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.8167 │ 1.2235 │ 66.75 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.8133 │ 1.2235 │ 66.48 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.7481 │ 1.8547 │ 40.33 % │
│ cpac_sub-0010042_ses-1.WM_63.seg_tissue… │ 0.6710 │ 1.7051 │ 39.35 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.7274 │ 1.8547 │ 39.22 % │
└──────────────────────────────────────────┴───────────┴───────────┴───────────┘
[6]:
cpac parse-resources \
--filter_field efficiency \
--filter_group highest \
--filter_count 5 \
cpac_runs/enough/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┓
┃ ┃ Memory ┃ Memory ┃ Memory ┃
┃ Task ID ┃ Used ┃ Estimated ┃ Efficien… ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━┩
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.7283 │ 1.2235 │ 59.53 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.7850 │ 1.8547 │ 42.32 % │
│ cpac_sub-0010042_ses-1.CSF_64.seg_tissu… │ 0.7159 │ 1.7051 │ 41.99 % │
│ cpac_sub-0010042_ses-1.WM_64.seg_tissue… │ 0.6658 │ 1.7051 │ 39.05 % │
│ cpac_sub-0010042_ses-1.GM_64.seg_tissue… │ 0.6486 │ 1.7051 │ 38.04 % │
└──────────────────────────────────────────┴───────────┴───────────┴───────────┘
Let’s look at the other bottom five efficiency and the top and bottom five for each of the other fields for one of these callback.logs.
[7]:
cpac parse-resources \
--filter_field efficiency \
--filter_group lowest \
--filter_count 5 \
cpac_runs/enough/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┓
┃ ┃ Memory ┃ Memory ┃ Memory ┃
┃ Task ID ┃ Used ┃ Estimated ┃ Efficien… ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━┩
│ cpac_sub-0010042_ses-1.montage_mni_anat… │ 0.2135 │ 13.8000 │ 1.55 % │
│ cpac_sub-0010042_ses-1.anat_reorient_0 │ 0.2233 │ 13.8000 │ 1.62 % │
│ cpac_sub-0010042_ses-1.qc_skullstrip_63… │ 0.2272 │ 13.8000 │ 1.65 % │
│ cpac_sub-0010042_ses-1.qc_skullstrip_63… │ 0.2272 │ 13.8000 │ 1.65 % │
│ cpac_sub-0010042_ses-1.qc_skullstrip_63… │ 0.2272 │ 13.8000 │ 1.65 % │
└──────────────────────────────────────────┴───────────┴───────────┴───────────┘
[8]:
cpac parse-resources \
--filter_field estimate \
--filter_group highest \
--filter_count 5 \
cpac_runs/enough/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┓
┃ ┃ Memory ┃ Memory ┃ Memory ┃
┃ Task ID ┃ Used ┃ Estimated ┃ Efficien… ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━┩
│ cpac_sub-0010042_ses-1.montage_mni_anat… │ 0.2135 │ 13.8000 │ 1.55 % │
│ cpac_sub-0010042_ses-1.montage_mni_anat… │ 0.2279 │ 13.8000 │ 1.65 % │
│ cpac_sub-0010042_ses-1.anat_reorient_0 │ 0.2233 │ 13.8000 │ 1.62 % │
│ cpac_sub-0010042_ses-1.montage_mni_anat… │ 0.2279 │ 13.8000 │ 1.65 % │
│ cpac_sub-0010042_ses-1.qc_skullstrip_82… │ 0.2348 │ 13.8000 │ 1.70 % │
└──────────────────────────────────────────┴───────────┴───────────┴───────────┘
[9]:
cpac parse-resources \
--filter_field runtime \
--filter_group highest \
--filter_count 5 \
cpac_runs/enough/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┓
┃ ┃ Memory ┃ Memory ┃ Memory ┃
┃ Task ID ┃ Used ┃ Estimated ┃ Efficien… ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━┩
│ cpac_sub-0010042_ses-1.segment_64 │ 1.1257 │ 3.4958 │ 32.20 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.9344 │ 3.0513 │ 30.62 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.7850 │ 1.8547 │ 42.32 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.7283 │ 1.2235 │ 59.53 % │
│ cpac_sub-0010042_ses-1.CSF_64.seg_tissu… │ 0.7159 │ 1.7051 │ 41.99 % │
└──────────────────────────────────────────┴───────────┴───────────┴───────────┘
[10]:
cpac parse-resources \
--filter_field estimate \
--filter_group lowest \
--filter_count 5 \
cpac_runs/enough/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┓
┃ ┃ Memory ┃ Memory ┃ Memory ┃
┃ Task ID ┃ Used ┃ Estimated ┃ Efficien… ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━┩
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.7283 │ 1.2235 │ 59.53 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.3601 │ 1.2806 │ 28.12 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.2212 │ 1.4200 │ 15.58 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.3732 │ 1.5000 │ 24.88 % │
│ cpac_sub-0010042_ses-1.CSF_64.seg_tissu… │ 0.7159 │ 1.7051 │ 41.99 % │
└──────────────────────────────────────────┴───────────┴───────────┴───────────┘
[11]:
cpac parse-resources \
--filter_field runtime \
--filter_group lowest \
--filter_count 5 \
cpac_runs/enough/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┓
┃ ┃ Memory ┃ Memory ┃ Memory ┃
┃ Task ID ┃ Used ┃ Estimated ┃ Efficien… ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━┩
│ cpac_sub-0010042_ses-1.CSF_64.seg_tissu… │ 0.1778 │ 2.5000 │ 7.11 % │
│ cpac_sub-0010042_ses-1.func_ingress_sub… │ 0.1788 │ 2.0000 │ 8.94 % │
│ cpac_sub-0010042_ses-1.func_ingress_sub… │ 0.1789 │ 2.0000 │ 8.95 % │
│ cpac_sub-0010042_ses-1.func_ingress_sub… │ 0.1789 │ 2.0000 │ 8.95 % │
│ cpac_sub-0010042_ses-1.func_ingress_sub… │ 0.1796 │ 2.0000 │ 8.98 % │
└──────────────────────────────────────────┴───────────┴───────────┴───────────┘
1.2. Memory optimized runs¶
Now that we have observed memory usage specific to our system + configuration + data shape, we can use these observations to inform the estimates of our subesquent runs. Here we’ll run another subject and re-run the same intial subject so we can compare the performance. We’ll use our original desired memory constraints.
In this example, we’ll use a buffer of 25% to demonstrate adjusting the buffer. If you don’t set a buffer, the default of 10% will be used. If you don’t want a buffer, you can set the buffer to 0.
We can do this with a pipeline configuration like
anat-only-optimized.yml
FROM: anat-only
pipeline_setup:
system_config:
observed_usage:
callback_log: cpac_runs/enough/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log
buffer: 25
or we can use commandline flags like
[12]:
cpac run \
s3://fcp-indi/data/Projects/ADHD200/RawDataBIDS/NYU cpac_runs/optimized participant \
--preconfig anat-only \
--participant_label 0010042 5971050 \
--n_cpus 4 \
--mem_gb 10 \
--runtime_usage cpac_runs/enough/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log \
--runtime_buffer 25
Loading 🐳 Docker
[…]
Skipping bids-validator for S3 datasets...
Loading the 'anat-only' pre-configured pipeline.
#### Running C-PAC for 0010042, 5971050
Number of participants to run in parallel: 1
Input directory: s3://fcp-indi/data/Projects/ADHD200/RawDataBIDS/NYU
Output directory: […]/cpac_runs/optimized/output
Working directory: […]/cpac_runs/optimized/working
Log directory: […]/cpac_runs/optimized/log
Remove working directory: False
Available memory: 10.0 (GB)
Available threads: 4
Number of threads for ANTs: 1
Parsing s3://fcp-indi/data/Projects/ADHD200/RawDataBIDS/NYU..
Connecting to AWS: fcp-indi anonymously...
gathering files from S3 bucket (s3.Bucket(name='fcp-indi')) for data/Projects/ADHD200/RawDataBIDS/NYU
Starting participant level processing
Run called with config file […]/cpac_runs/optimized/cpac_pipeline_config_2022-04-14T15-30-12Z.yml
220414-15:30:20,887 nipype.workflow INFO:
Run command: run s3://fcp-indi/data/Projects/ADHD200/RawDataBIDS/NYU cpac_runs/optimized participant --preconfig anat-only --participant_label 0010042 5971050 --n_cpus 4 --mem_gb 10 --runtime_usage cpac_runs/enough/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log --runtime_buffer 25
C-PAC version: 1.8.4.dev
Setting maximum number of cores per participant to 4
Setting number of participants at once to 1
Setting OMP_NUM_THREADS to 1
Setting MKL_NUM_THREADS to 1
Setting ANTS/ITK thread usage to 1
Maximum potential number of cores that might be used during this run: 4
[…]
The following nodes used excessive resources:
---------------------------------------------
cpac_sub-0010042_ses-1
.sinker_desc-brain_T1w_50
**memory_gb**
runtime > estimated
0.258327484375 > 0.23091316162109377
cpac_sub-0010042_ses-1
.sinker_label-GM_probseg_71
**memory_gb**
runtime > estimated
0.258304595703125 > 0.22700309814453123
cpac_sub-0010042_ses-1
.nii_space-template_desc-brain_T1w_57
**memory_gb**
runtime > estimated
0.25536346484375 > 0.22719383300781248
cpac_sub-0010042_ses-1
.nii_label-WM_desc-preproc_mask_78
**memory_gb**
runtime > estimated
0.236785888671875 > 0.22455692260742188
---------------------------------------------
220414-15:31:25,9 nipype.workflow INFO:
End of subject workflow cpac_sub-0010042_ses-1
CPAC run complete:
Pipeline configuration: cpac_anat
Subject workflow: cpac_sub-0010042_ses-1
[…]
220414-15:31:25,33 nipype.workflow INFO:
Run command: run s3://fcp-indi/data/Projects/ADHD200/RawDataBIDS/NYU cpac_runs/optimized participant --preconfig anat-only --participant_label 0010042 5971050 --n_cpus 4 --mem_gb 10 --runtime_usage cpac_runs/enough/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log --runtime_buffer 25
C-PAC version: 1.8.4.dev
Setting maximum number of cores per participant to 4
Setting number of participants at once to 1
Setting OMP_NUM_THREADS to 1
Setting MKL_NUM_THREADS to 1
Setting ANTS/ITK thread usage to 1
Maximum potential number of cores that might be used during this run: 4
[…]
The following nodes used excessive resources:
---------------------------------------------
cpac_sub-5971050_ses-1
.sinker_space-T1w_desc-brain_mask_47
**memory_gb**
runtime > estimated
0.239810943359375 > 0.23887157470703124
cpac_sub-5971050_ses-1
.sinker_desc-brain_T1w_50
**memory_gb**
runtime > estimated
0.23128891015625 > 0.23091316162109377
cpac_sub-5971050_ses-1
.nii_label-CSF_mask_73
**memory_gb**
runtime > estimated
0.23983383203125 > 0.22700309814453123
cpac_sub-5971050_ses-1
.sinker_label-GM_probseg_71
**memory_gb**
runtime > estimated
0.23983383203125 > 0.22700309814453123
cpac_sub-5971050_ses-1
.nii_space-template_desc-brain_T1w_57
**memory_gb**
runtime > estimated
0.2322998046875 > 0.22719383300781248
cpac_sub-5971050_ses-1
.ANTS_T1_to_template_51
.inverse_all_transform_flags
**memory_gb**
runtime > estimated
0.2548370361328125 > 0.2273988720703125
cpac_sub-5971050_ses-1
.nii_from-template_to-T1w_mode-image_xfm_59
**memory_gb**
runtime > estimated
0.255855560546875 > 0.22507190673828126
cpac_sub-5971050_ses-1
.sinker_from-template_to-T1w_mode-image_xfm_59
**memory_gb**
runtime > estimated
0.255855560546875 > 0.22446632324218752
---------------------------------------------
220414-15:32:37,121 nipype.workflow INFO:
End of subject workflow cpac_sub-5971050_ses-1
CPAC run complete:
Pipeline configuration: cpac_anat
Subject workflow: cpac_sub-5971050_ses-1
[…]
Now let’s see how our efficiency compares across runs.
[13]:
cpac parse-resources \
--filter_field efficiency \
--filter_group highest \
--filter_count 5 \
cpac_runs/optimized/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┓
┃ ┃ Memory ┃ Memory ┃ Memory ┃
┃ Task ID ┃ Used ┃ Estimated ┃ Efficien… ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━┩
│ cpac_sub-0010042_ses-1.sinker_label-GM_… │ 0.2583 │ 0.2270 │ 113.79 % │
│ cpac_sub-0010042_ses-1.nii_space-templa… │ 0.2554 │ 0.2272 │ 112.40 % │
│ cpac_sub-0010042_ses-1.sinker_desc-brai… │ 0.2583 │ 0.2309 │ 111.87 % │
│ cpac_sub-0010042_ses-1.nii_label-WM_des… │ 0.2368 │ 0.2246 │ 105.45 % │
│ cpac_sub-0010042_ses-1.montage_mni_anat… │ 0.3656 │ 0.3714 │ 98.45 % │
└──────────────────────────────────────────┴───────────┴───────────┴───────────┘
[14]:
cpac parse-resources \
--filter_field efficiency \
--filter_group lowest \
--filter_count 5 \
cpac_runs/optimized/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┓
┃ ┃ Memory ┃ Memory ┃ Memory ┃
┃ Task ID ┃ Used ┃ Estimated ┃ Efficien… ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━┩
│ cpac_sub-0010042_ses-1.resampled_T1w-br… │ 0.1883 │ 2.0000 │ 9.41 % │
│ cpac_sub-0010042_ses-1.montage_mni_anat… │ 0.2606 │ 0.5124 │ 50.87 % │
│ cpac_sub-0010042_ses-1.CSF_64.seg_tissu… │ 0.1830 │ 0.3170 │ 57.74 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.1926 │ 0.3283 │ 58.65 % │
│ cpac_sub-0010042_ses-1.get_pve_wm_64 │ 0.2566 │ 0.4225 │ 60.73 % │
└──────────────────────────────────────────┴───────────┴───────────┴───────────┘
[15]:
cpac parse-resources \
--filter_field estimate \
--filter_group highest \
--filter_count 5 \
cpac_runs/optimized/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┓
┃ ┃ Memory ┃ Memory ┃ Memory ┃
┃ Task ID ┃ Used ┃ Estimated ┃ Efficien… ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━┩
│ cpac_sub-0010042_ses-1.resampled_T1w-br… │ 0.1883 │ 2.0000 │ 9.41 % │
│ cpac_sub-0010042_ses-1.segment_64 │ 1.1376 │ 1.4072 │ 80.84 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.9533 │ 1.1681 │ 81.62 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.7189 │ 0.9812 │ 73.27 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.7630 │ 0.9104 │ 83.81 % │
└──────────────────────────────────────────┴───────────┴───────────┴───────────┘
[16]:
cpac parse-resources \
--filter_field runtime \
--filter_group highest \
--filter_count 5 \
cpac_runs/optimized/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┓
┃ ┃ Memory ┃ Memory ┃ Memory ┃
┃ Task ID ┃ Used ┃ Estimated ┃ Efficien… ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━┩
│ cpac_sub-0010042_ses-1.segment_64 │ 1.1376 │ 1.4072 │ 80.84 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.9533 │ 1.1681 │ 81.62 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.7630 │ 0.9104 │ 83.81 % │
│ cpac_sub-0010042_ses-1.ANTS_T1_to_templ… │ 0.7189 │ 0.9812 │ 73.27 % │
│ cpac_sub-0010042_ses-1.WM_64.seg_tissue… │ 0.6409 │ 0.8323 │ 77.00 % │
└──────────────────────────────────────────┴───────────┴───────────┴───────────┘
[17]:
cpac parse-resources \
--filter_field estimate \
--filter_group lowest \
--filter_count 5 \
cpac_runs/optimized/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┓
┃ ┃ Memory ┃ Memory ┃ Memory ┃
┃ Task ID ┃ Used ┃ Estimated ┃ Efficien… ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━┩
│ cpac_sub-0010042_ses-1.CSF_64.seg_tissu… │ 0.1800 │ 0.2223 │ 80.96 % │
│ cpac_sub-0010042_ses-1.func_ingress_sub… │ 0.1815 │ 0.2245 │ 80.84 % │
│ cpac_sub-0010042_ses-1.sinker_from-temp… │ 0.2025 │ 0.2245 │ 90.24 % │
│ cpac_sub-0010042_ses-1.func_ingress_sub… │ 0.1815 │ 0.2245 │ 80.84 % │
│ cpac_sub-0010042_ses-1.func_ingress_sub… │ 0.1800 │ 0.2245 │ 80.17 % │
└──────────────────────────────────────────┴───────────┴───────────┴───────────┘
[18]:
cpac parse-resources \
--filter_field runtime \
--filter_group lowest \
--filter_count 5 \
cpac_runs/optimized/log/pipeline_cpac_anat/sub-0010042_ses-1/callback.log
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┳━━━━━━━━━━━┓
┃ ┃ Memory ┃ Memory ┃ Memory ┃
┃ Task ID ┃ Used ┃ Estimated ┃ Efficien… ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━╇━━━━━━━━━━━┩
│ cpac_sub-0010042_ses-1.id_string_from-T… │ 0.1762 │ 0.2311 │ 76.28 % │
│ cpac_sub-0010042_ses-1.json_desc-brain_… │ 0.1762 │ 0.2311 │ 76.28 % │
│ cpac_sub-0010042_ses-1.id_string_space-… │ 0.1762 │ 0.2343 │ 75.22 % │
│ cpac_sub-0010042_ses-1.json_desc-reorie… │ 0.1762 │ 0.2257 │ 78.08 % │
│ cpac_sub-0010042_ses-1.id_string_from-t… │ 0.1762 │ 0.2344 │ 75.18 % │
└──────────────────────────────────────────┴───────────┴───────────┴───────────┘
Here are some box-and-whisker plots (generated in visualize_observed_usage.ipynb comparing the efficiency, estimates, and observed memory usage of the four runs we completed above:
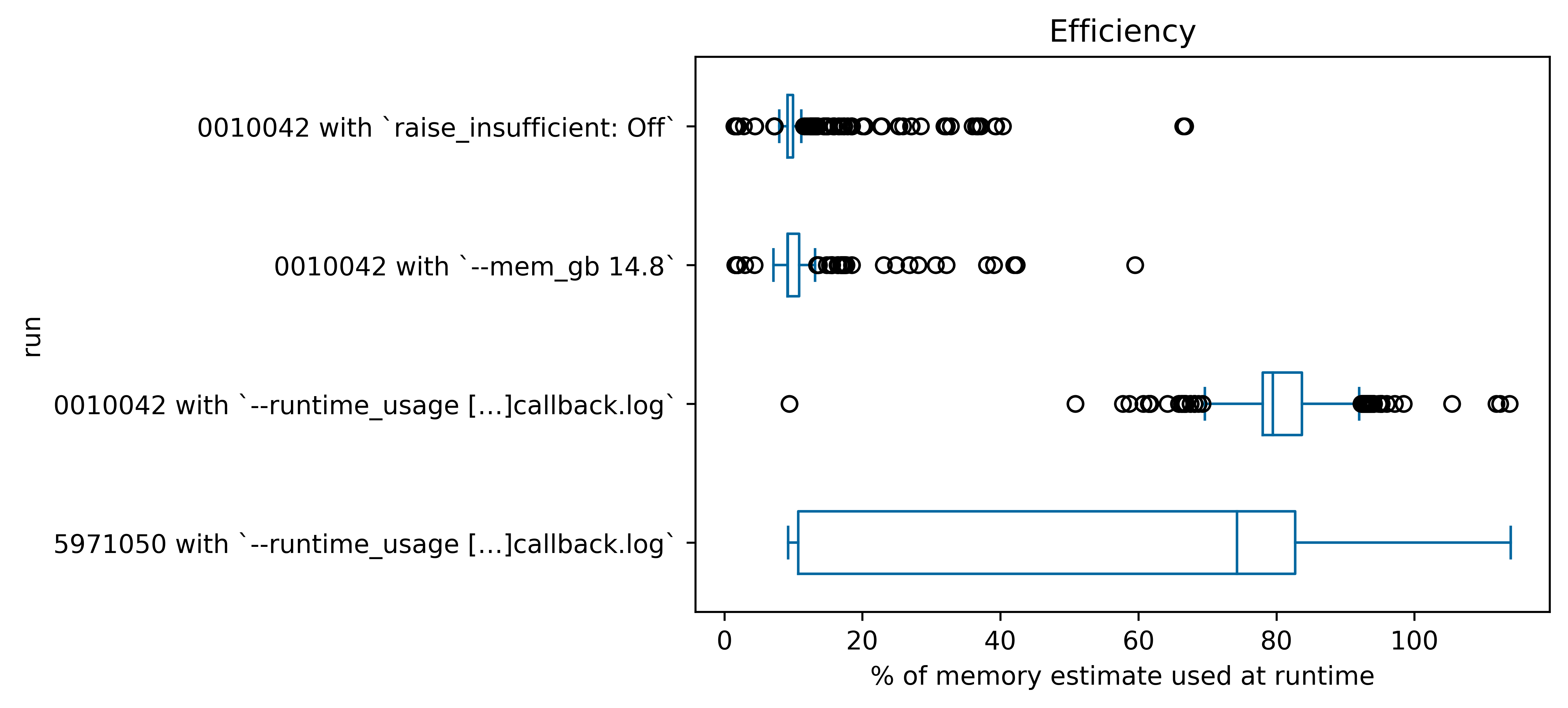
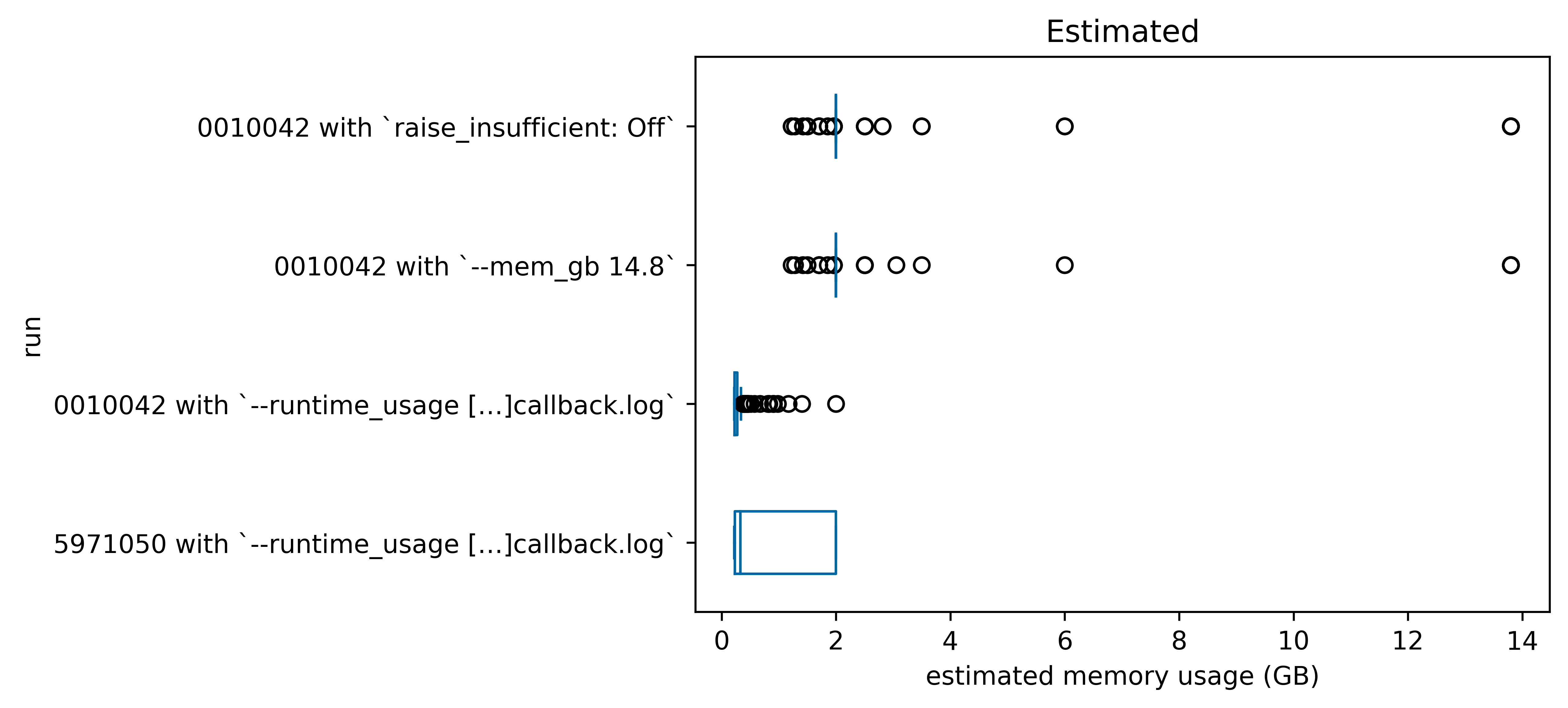
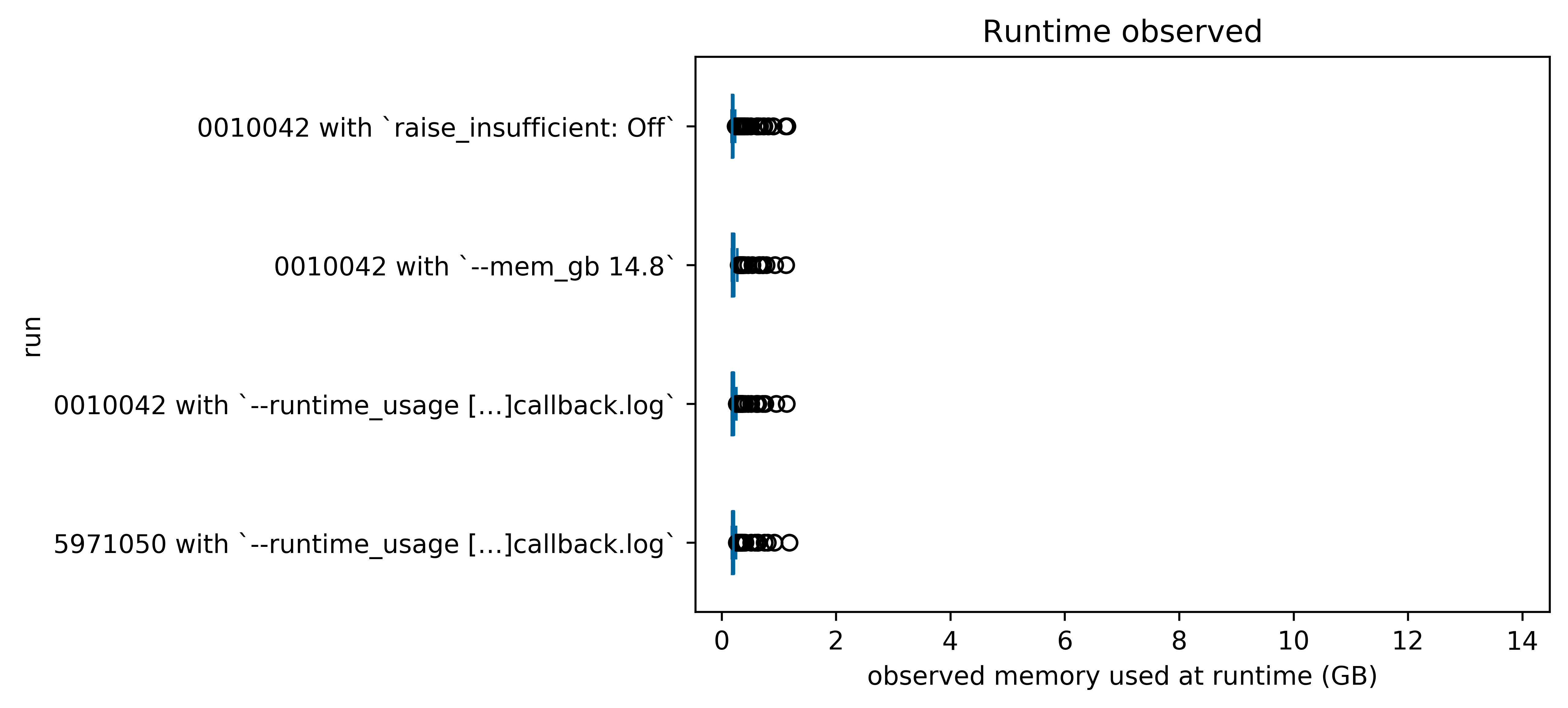
1.3. Notes¶
1.3.1. Total memory usage¶
These estimates and observations are per-node. At any given moment, you’ll have up to n_cpus nodes running at once, plus C-PAC and Nipype will be using some memory, so a worst-case scenario for memory usage is approximately the heaviest n_cpus nodes + 1 GB for overhead.
In this example, we set n_cpus to 4. For the optimized re-run in this example, the four greediest nodes + 1 GB for overhead = (1.1376 + 0.9533 + 0.7630 + 0.7189 + 1) GB = 4.5728 GB, which is well within the specified limit of 10 GB. In most cases, the greediest nodes won’t be peaking simultaneously, so we won’t expect an out-of-memory error on an optimized run, but they are possible, even if no individual node is overruning by much.