After Warping Options¶
Spatial Smoothing¶
The main goal of smoothing is to increase the signal-to-noise ratio of an image. This is done by removing high-frequency information (assumed to consist mostly of noise) while preserving signals on larger spatial scales (those which stretch across multiple voxels and are most likely to reflect neuronal activity). Smoothing can also help reduce the effect of anatomical variability when comparing activations between subjects (Poldrack et al., 2011). Additionally, smoothing with a Gaussian kernel (described below) will cause the distribution of signals found in an image to more closely match the assumptions of many statistical tests often used on fMRI data.
Computation and Analysis Considerations¶
Smoothing is accomplished by applying a Gaussian filter (also known as a kernel) to the original image. This results in the signal of each voxel being replaced by the weighted average of its neighbors (Ashby, 2011). It is the shape of the kernel that defines the weights applied during smoothing. As a Gaussian kernel is essentially a normal distribution, weight is always strongest at the voxel being smoothed and decreases with distance at a rate that depends on the width of the distribution. Wider distributions result in greater smoothing, as more distant voxels recieve higher weights than they would with a thinner kernel. Unlike in statistics, where the width of a distribution is described by the standard deviation, the shape of a smoothing kernel is usually described by the width of the distribution at half of its maximum value. This is known as the Full Width Half Maximum (FWHM). The figure below shows the smoothing effect of different FWHM values.
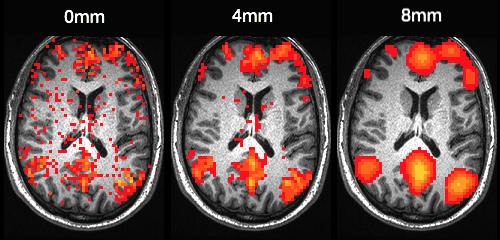
Note: In C-PAC, smoothing is applied after calculating individual-level analyses (except for VMHC, for which smoothing is applied prior to calculation). This preserves the structure of the data as much as possible prior to statistical analysis.
Applications and Guidelines¶
The amount of smoothing applied should be the minimum necessary to achive the desired result. As a general rule, a FWHM of twice the voxel dimension is a good starting point (Poldrack et al., 2011). That being said, different situations will require adjustment of this value. In particular, the FWHM should never be larger than the smallest activation pattern you are interested in detecting; any larger value may result in signal loss, either by smoothing away very small signals, or smoothing two independent signals into one (Ashby, 2011).
Care must be taken when utilizing methods such as Regional Homogeneity (ReHo) and Multi-voxel Pattern Analysis (MVPA) which are sensitive to the activation of individual voxels, as smoothing prior to analysis may bias results by introducing artificial spatial correlation. In these cases, if smoothing is desired, such as in order to correct for multiple comparisons using Gaussian Random Field Theory, it must be applied after the fact.
Z-Scored Derivatives¶
C-PAC can take all of the outputs it produces and standardize their values as z-scores.
Configuring C-PAC to Run After-Warping Steps¶
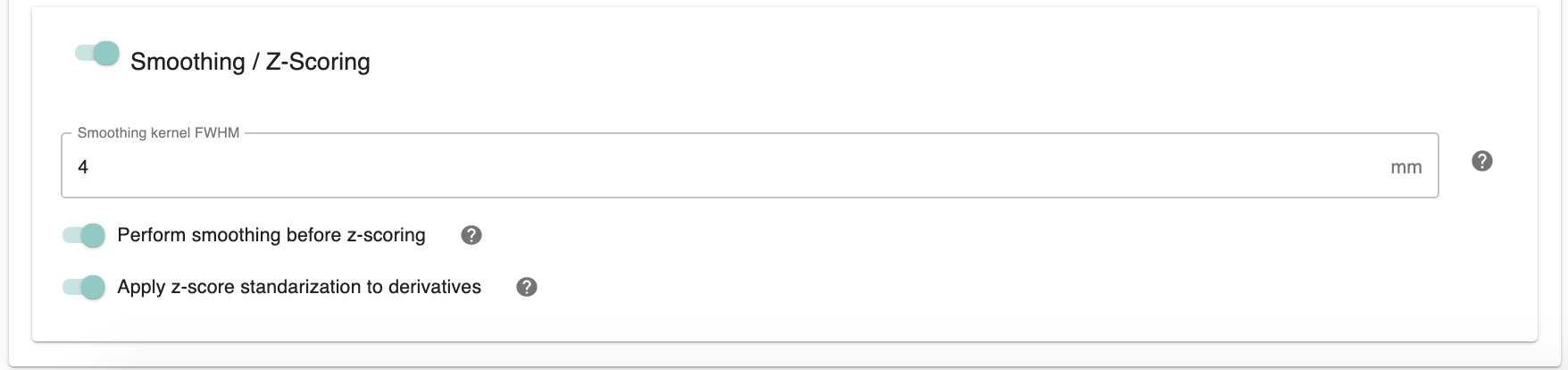
Smoothing / Z-Scoring: Smooth the derivative outputs. On - Run smoothing and output only the smoothed outputs. On/Off - Run smoothing and output both the smoothed and non-smoothed outputs. Off - Don’t run smoothing.
Smoothing Kernel FWHM (in mm) [integer/decimal]: Full Width at Half Maximum of the Gaussian kernel used during spatial smoothing. Can be a single value or multiple values separated by commas. Note that spatial smoothing is run as the last step in the individual-level analysis pipeline, such that all derivatives are output both smoothed and unsmoothed.
Perfom smoothing before z-Scoring [On, Off]: Choose whether to smooth before or after z-scoring.
Apply z-score Standardize to derivatives [On, Off]: z-score standardize the derivatives. This is required for group-level analysis. On - Run z-scoring and output only the z-scored outputs. On/Off - Run z-scoring and output both the z-scored and raw score versions of the outputs. Off - Don’t run z-scoring.
Configuration Using a YAML File¶
The following nested key/value pairs that will be set to these defaults if not defined in your pipeline configuration YAML:
# OUTPUTS AND DERIVATIVES
# -----------------------
post_processing:
spatial_smoothing:
run: On
# Smooth the derivative outputs.
# Set as ['nonsmoothed'] to disable smoothing. Set as ['smoothed', 'nonsmoothed'] to get both.
#
# Options:
# ['smoothed', 'nonsmoothed']
output: ['smoothed']
# Tool to use for smoothing.
# 'FSL' for FSL MultiImageMaths for FWHM provided
# 'AFNI' for AFNI 3dBlurToFWHM for FWHM provided
smoothing_method: ['FSL']
# Full Width at Half Maximum of the Gaussian kernel used during spatial smoothing.
# this is a fork point
# i.e. multiple kernels - fwhm: [4,6,8]
fwhm: [4]
z-scoring:
run: On
# z-score standardize the derivatives. This may be needed for group-level analysis.
# Set as ['raw'] to disable z-scoring. Set as ['z-scored', 'raw'] to get both.
#
# Options:
# ['z-scored', 'raw']
output: ['z-scored']
External Resources¶
References¶
Ashby, F.G., (2011). Preprocessing. Statistical Analysis of MRI Data. Cambridge, MA: MIT Press.
Poldrack, R. A., Mumford, J., & Nichols, T. (2011). Preprocessing fMRI data. In Handbook of Functional MRI Data Analysis. Cambridge: Cambridge University Press.