FSL-FEAT/Randomise Group Analysis¶
Overview¶
C-PAC uses the FSL/FEAT tool to compare findings across groups. You can construct models using a participant list and a phenotype file, select derivatives to be predicted by the model, and define contrasts between conditions using a custom CSV file. Then FSL/FEAT will run a second-level General Linear Model (GLM) for you.
In addition, FMRIB’s FSL Randomise package is also available in C-PAC for the execution of non-parametric permutation inference. FSL-Randomise can use the same group models you generate or build/edit via the C-PAC model builder for FSL-FEAT.
There are two ways to set up FSL-FEAT/Randomise group-level analysis for C-PAC:
The FLAME Model Presets: this allows you to generate a pre-configured analysis model.
The Group Analysis Model Builder: this allows you to specify a model from scratch, or to modify any of the generated presets mentioned above.
The following links provide an introduction to how groups are compared using FSL, as well as how to define contrasts:
Group Analysis FSL-FEAT/Randomise Presets¶
C-PAC has a selection of model presets designed to run commonly-used group analysis designs for FSL-FEAT/FLAME. These correspond to examples provided on FSL’s user guide for FEAT/FLAME. The preset generator will create a group analysis YAML configuration file that you can plug directly into C-PAC and run.
The presets that are generated:
are meant to get you up and running quickly
are in the form of a group analysis configuration YAML file that can be plugged directly into C-PAC and run
include a complete design matrix as an input, which you can view before running
include already-configured contrasts in a custom contrasts .CSV file, which can be edited before running
can be modified to your liking either by using the standard Group Analysis Model Builder, or by hand via text editor
are all in one place in the output directory you specify in the Preset Generator
Preset options: Currently, there are 5 presets to choose from. More are on their way. If you have any commonly-used or useful group model designs you’d like to see as a preset, please let us know on GitHub or Neurostars!
The available presets are:
From Terminal¶
You can generate any of these presets using the C-PAC command-line interface (CLI):
cpac group feat load-preset <preset type>
Enter any of the following in place of <preset type> for the type of analysis you want to run:
single_grp_avg - Single Group Average (One-Sample T-Test)
single_grp_cov - Single Group Average with Additional Covariate
unpaired_two - Unpaired Two-Group Difference (Two-Sample Unpaired T-Test)
paired_two - Paired Two-Group Difference (Two-Sample Paired T-Test)
tripled_two - Tripled Two-Group Difference (‘Tripled’ T-Test)
You can get more information about the required inputs for each preset with the --help flag. For example, to check the parameters for a two-sample unpaired t-test:
cpac group feat load-preset unpaired-two --help
This will produce:
Usage: run.py group feat load-preset unpaired-two [OPTIONS] PIPELINE_DIR
Z_THRESH P_THRESH PHENO_FILE
PHENO_SUB COVARIATE
MODEL_NAME
Options:
--output-dir, --output_dir TEXT
--group-participants, --group_participants TEXT
--help Show this message and exit.
Following this, you could generate a ready-to-run two-sample unpaired t-test by running the following, assuming the phenotype CSV has a column of participant IDs named “subject_id” and a column named “diagnosis”, which is the covariate you wish to test:
cpac group feat load-preset unpaired-two /path/to/group_participant_list.txt 2.3 0.05
/path/to/phenotypic_file.csv subject_id diagnosis grp_analysis1
--output-dir /path/to/output_dir
You will receive a message like this shortly after:
Group-level analysis participant list written:
/path/to/output_dir/group_analysis_participants.txt
CSV file written:
/path/to/output_dir/cpac_group_analysis/grp_analysis1/design_matrix_grp_analysis1.csv
CSV file written:
/path/to/output_dir/cpac_group_analysis/grp_analysis1/contrasts_matrix_grp_analysis1.csv
Group-level analysis configuration YAML file written:
/path/to/output_dir/cpac_group_analysis/grp_analysis1/group_config_grp_analysis1.yml
The message indicates that all of the associated files have been generated. Most importantly, it generated a group configuration file that you can use to build and run your model. Once complete, you can review the model generated to make sure it meets your expectations. If you wish to tweak the model generated by a preset, you can load the group configuration file into the model builder (more details below) and make any modifications necessary.
When you are happy with your model, you can go ahead to build the model for each of your selected derivatives:
cpac group feat build-models /path/to/group_config.yml
Once this is complete, a design matrix will have been generated for each derivative, session, scan, and nuisance regression strategy you had in your pipeline directory. These are available for review in the group-level analysis output directory you specified in your group configuration file (or during preset generation). In addition, a contrasts.csv file will be generated in the top level of the FSL group model directory (in your selected group-level analysis output directory). This contrasts file will be set up in accordance with the design matrices created for your pipeline outputs, and will look like this:

The first column should be filled with labels for the contrasts that you can define - these do not have to follow any particular convention, and can be whatever works best for your experiment. The remainder of the cells can be populated with contrast weights according to your needs.
If you would like to add f-tests, add each f-test as a column to the CSV with the label f_test_# and the assign weights to each contrast to be included in the f-test.
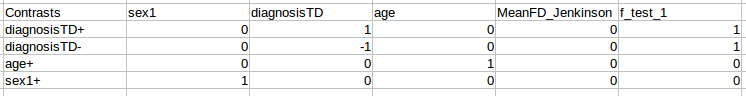
Once you have specified your contrasts, you can start FSL FEAT or Randomise with the commands:
cpac group feat run /path/to/group_config.yml
cpac group feat randomise /path/to/group_config.yml
C-PAC Group Analysis Model Builder¶
Use the model builder to create an FSL FEAT/Randomise model from scratch, or to modify any of the generated presets.
From the terminal¶
Similar to the pipeline configuration YAML file, the group configuration YAML file allows you to configure your runs with key-value combinations. From terminal, you can quickly generate a default group configuration YAML file template in the directory you are in:
cpac utils group-config new-template
This will generate a group configuration file that you can then modify to make your selections. See below:
# General Group-Level Analysis Settings
##############################################################################
# The main input of group-level analysis- the output directory of your individual-level analysis pipeline run (pre-processing & derivatives for each participant). This should be a path to your C-PAC individual-level run's pipeline folder, which includes the sub-directories labeled with the participant IDs.
pipeline_dir: /path/to/output_dir
# (Optional) Full path to a list of participants to be included in the model. You can use this to easily prune participants from your model. In group-level analyses involving phenotype files, this allows you to prune participants without removing them from the phenotype CSV/TSV file. This should be a text file with one subject per line. An easy way to manually create this file is to copy the participant ID column from your phenotype file.
participant_list: None
# Full path to the directory where CPAC should place group-level analysis outputs and any applicable statistical model files.
output_dir: /path/to/output/dir
#Much like the working directory for individual-level analysis, this is where the intermediate and working files will be stored during your run. This directory can be deleted later on. However, saving this directory allows the group analysis run to skip steps that have been already completed, in the case of re-runs.
work_dir: /path/to/work/dir
#Where to write out log information for your group analysis run.
log_dir: /path/to/log/dir
# The path to your FSL installation directory. This can be left as 'FSLDIR' to grab your system's default FSL installation. However, if you prefer to use a specific install of FSL, you can enter the path here.
FSLDIR: FSLDIR
# FSL-FEAT
##############################################################################
# Run FSL FEAT group-level analysis.
run_fsl_feat : [1]
# How many statistical models to run in parallel. This number depends on computing resources.
num_models_at_once : 1
# Specify a name for the new model.
model_name: model_name_here
# Phenotype file
# Full path to a .csv or .tsv file containing EV/regressor information for each subject.
pheno_file: /path/to/phenotypic/file.csv
# Name of the participants column in your phenotype file.
participant_id_label: Participant
# Specify which EVs from your phenotype are categorical or numerical. Of those which are numerical, specify which are to be demeaned.
# ev_selections: {'demean': ['Age'], 'categorical': ['Sex', 'Diagnosis']}
ev_selections: {'demean': [], 'categorical': []}
# Specify the formula to describe your model design. Essentially, including EVs in this formula inserts them into the model. The most basic format to include each EV you select would be 'EV + EV + EV + ..', etc. You can also select to include MeanFD, Measure_Mean, and Custom_ROI_Mean here. See the C-PAC User Guide for more detailed information regarding formatting your design formula.
# design_formula: Sex + Diagnosis + Age + MeanFD_Jenkinson + Custom_ROI_Mean
design_formula:
# Choose the derivatives to run the group model on.
#
# These must be written out as a list, and must be one of the options listed below.
#
# For z-scored analyses:
# 'alff_to_standard_zstd', 'alff_to_standard_smooth_zstd', 'falff_to_standard_zstd', 'falff_to_standard_smooth_zstd', 'reho_to_standard_zstd', 'reho_to_standard_smooth_zstd', 'sca_roi_files_to_standard_fisher_zstd', 'sca_roi_files_to_standard_smooth_fisher_zstd', 'vmhc_fisher_zstd_zstat_map', 'dr_tempreg_maps_zstat_files_to_standard', 'dr_tempreg_maps_zstat_files_to_standard_smooth', 'sca_tempreg_maps_zstat_files', 'sca_tempreg_maps_zstat_files_smooth', 'centrality_outputs_zstd', 'centrality_outputs_smoothed_zstd'
#
# Example input: derivative_list : ['alff_to_standard_smooth_zstd', 'sca_roi_files_to_standard_smooth_fisher_zstd']
#
derivative_list: []
# Choose whether to use a group mask or individual-specific mask when calculating the output means to be used as a regressor.
#
# This only takes effect if you include the 'Measure_Mean' regressor in your Design Matrix Formula.
mean_mask: ['Group Mask']
# Full path to a NIFTI file containing one or more ROI masks. The means of the masked regions will then be computed for each subject's output and will be included in the model as regressors (one for each ROI in the mask file) if you include 'Custom_ROI_Mean' in the Design Matrix Formula.
# custom_roi_mask: /path/to/mask.nii.gz
custom_roi_mask: None
# Choose the coding scheme to use when generating your model. 'Treatment' encoding is generally considered the typical scheme. Consult the User Guide for more information.
#
# Available options:
# 'Treatment', 'Sum'
#
coding_scheme: ['Treatment']
# Specify whether FSL should model the variance for each group separately.
#
# If this option is enabled, you must specify a grouping variable below.
group_sep: Off
# The name of the EV that should be used to group subjects when modeling variances.
#
# If you do not wish to model group variances separately, set this value to None.
grouping_var: None
# Only voxels with a Z-score higher than this value will be considered significant.
z_threshold: ['2.3']
# Significance threshold (P-value) to use when doing cluster correction for multiple comparisons.
p_threshold: ['0.05']
# For repeated measures only. Enter the session names in your dataset that you wish to include within the same model (this is for repeated measures / within-subject designs).\n\nTip: These will be the names listed as "unique_id" in the original individual-level participant list, or the labels in the original data directories you marked as {session} while creating the CPAC participant list.
# sessions_list: ['ses-01', 'ses-02']
sessions_list: []
# For repeated measures only. Enter the series names in your dataset that you wish to include within the same model (this is for repeated measures / within-subject designs).\n\nTip: These will be the labels listed under "func:" in the original individual-level participant list, or the labels in the original data directories you marked as {series} while creating the CPAC participant list.
# series_list: ['task-rest_run-1', 'task-rest_run-2']
series_list: []
# Specify your contrasts here. For example, if two of your available contrasts are EV1 and EV0, you can enter contrast descriptions such as 'EV1 - EV0 = 0' or 'EV1 = 0'. Consult the User Guide for more information about describing contrasts. Alternatively, you can provide your own custom-written contrasts matrix in a CSV file in the 'Custom Contrasts Matrix' field below.
# contrasts: ['C(Diagnosis)[T.ADHD] - C(Diagnosis)[T.Typical] = 0', 'C(Diagnosis)[T.Typical] - C(Diagnosis)[T.ADHD] = 0']
contrasts: []
# Optional: A list of f-test strings containing contrasts. If you do not wish to run f-tests, leave this blank.
f_tests: []
# Optional: Full path to a CSV file which specifies the contrasts you wish to run in group analysis. Consult the User Guide for proper formatting.
# If you wish to use the standard contrast builder, leave this field blank. If you provide a path for this option, CPAC will use your custom contrasts matrix instead, and will use the f-tests described in this custom file only (ignoring those you have input in the f-tests field above).
# If you wish to include f-tests, create a new column in your CSV file for each f-test named 'f_test_1', 'f_test_2', .. etc. Then, mark the contrasts you would like to include in each f-test with a 1, and mark the rest 0. Note that you must select at least two contrasts per f-test.
custom_contrasts: None
# FSL-Randomise
##############################################################################
# Run Randomise
run_randomise : [0]
# Number of permutations you would like to use when building up the null distribution to test against.
randomise_permutation : 500
# Cluster-based thresholding corrected for multiple comparisons by using the null distribution of the max (across the image) cluster mask.
randomise_thresh : 5
# Demean data temporally before model fitting.
randomise_demean : True
# From the FMRIB FSL-Randomise user guide: TFCE (Threshold-Free Cluster Enhancement) is a new method for finding 'clusters' in your data without having to define clusters in a binary way. Cluster-like structures are enhanced but the image remains fundamentally voxelwise.
randomise_tfce : True
When you are done specifying your model, you can go ahead to build the model for each of your selected derivatives:
cpac group feat build-models /path/to/group_config.yml
Once this is complete, a design matrix will have been generated for each derivative, session, scan, and nuisance regression strategy you had in your pipeline directory. These are available for review in the group-level analysis output directory you specified in your group configuration file (or during preset generation). In addition, a contrasts.csv file will be generated in the top level of the FSL group model directory (in your selected group-level analysis output directory). This contrasts file will be set up in accordance with the design matrices created for your pipeline outputs, and will look like this:

The first column should be filled with labels for the contrasts that you can define - these do not have to follow any particular convention, and can be whatever works best for your experiment. The remainder of the cells can be populated with contrast weights according to your needs.
If you would like to add f-tests, add each f-test as a column to the CSV with the label f_test_# and the assign weights to each contrast to be included in the f-test.
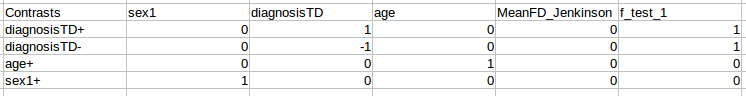
Once you have specified your contrasts, you can start FSL FEAT or Randomise with the commands:
cpac group feat run /path/to/group_config.yml
cpac group feat randomise /path/to/group_config.yml