Regional Homogeneity¶
Introduction & Background¶
Regional Homogeneity (ReHo) is a voxel-based measure of brain activity which evaluates the similarity or synchronization between the time series of a given voxel and its nearest neighbors (Zang et al., 2004). This measure is based on the hypothesis that intrinsic brain activity is manifested by clusters of voxels rather than single voxels. Kendall’s coefficient of concordance (KCC) (Kendall and Gibbons, 1990) is used as an index to evaluate the similarity of the time series within a cluster of a given voxel and its nearest neighbors. ReHo requires no a priori definition of ROIs and can provide information about the local/regional activity of regions throughout the brain.
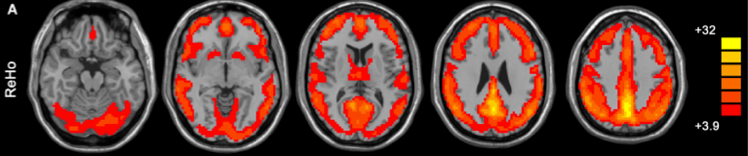
The figure above (taken from Yan and Zang, 2010) shows the default mode network as detected by ReHo analysis (colors indicate t values).
Computation and Analysis Considerations¶
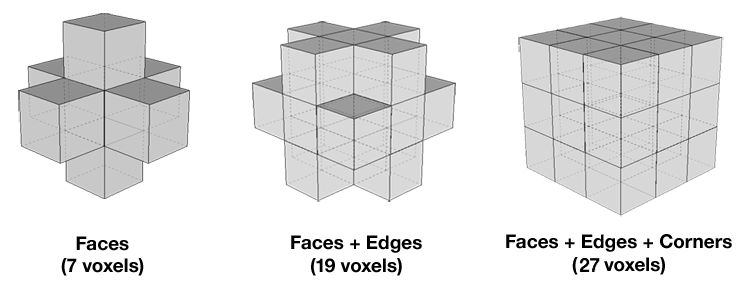
KCC is computed for every voxel in a subject, and is based on the time series of each voxel, the number of time points within a time series, and the number of voxels within a cluster (Zang et al, 2004). Depending on whether neighboring voxels are taken to include those on the side, edge, or corner of a given voxel, cluster size can be 7, 19, or 27 voxels, respectively. Values of KCC range from 0 to 1, with higher values indicating greater similarity between the activation pattern of a given voxel and that of its neighbors. Voxel-based maps are generated based on KCC values and then standardized using Z-scores in order to perform group analysis. For more detail on how CPAC handles these computations, please see the ReHo section of the developer documentation.
The results of ReHo analysis have been shown to vary significantly depending on both the number of neighbors in a cluster and the amount of spatial smoothing applied to the data. For example, Zang and colleagues (2004) found more neighbors and greater smoothing to yield greater differences between conditions in a motor task.
Applications and Recommendations¶
ReHo as an index of ongoing activity has been widely used in the resting-state literature. ReHo in the Default Mode Network (DMN) has been observed to increase during rest (Long et al., 2008) and decrease during task engagement (Zang et al., 2004), findings that fit well with the existing literature. In healthy subjects, ReHo measures are associated with individual differences in behavioral inhibition (Tian et al., 2012) and normal aging (Wu et al., 2007). In patient populations, altered ReHo has been observed in multiple conditions, including Alzheimer’s Disease (Liu et al., 2008), ADHD (Cao et al., 2006), and schizophrenia (Liu et al., 2006).
ReHo analysis can also be applied to task evoked fMRI data and is appropriate for application in block-based and slow event-related designs, but may not provide reliable results in rapid event-related studies. In such paradigms, the hemodynamic signal for a given timepoint is the result of multiple trials, and ReHo is unable to account for this overlapping of signals.
Configuring CPAC to Run ReHo¶
Regional Homogeneity (ReHo) - [Off, On]: Calculate Regional Homogeneity (ReHo) for all voxels.
Voxel Cluster Size - [7,19,27]: Number of neighboring voxels used when calculating ReHo. 7 (Faces), 19 (Faces + Edges), or 27 (Faces + Edges + Corners).
Configuration Without the GUI¶
The following nested key/value pairs will be set to these defaults if not defined in your pipeline configuration YAML.
regional_homogeneity:
# ReHo
# Calculate Regional Homogeneity (ReHo) for all voxels.
run: On
# space: Template or Native
target_space: ['Native']
# Number of neighboring voxels used when calculating ReHo
# 7 (Faces)
# 19 (Faces + Edges)
# 27 (Faces + Edges + Corners)
cluster_size: 27
Surface-based ReHo¶
New in version 1.8.7.
C-PAC now offers the computation of surface-based ReHo. This can be configured under surface_analysis in the pipeline file.
surface_analysis:
# Run freesurfer_abcd_preproc to obtain preprocessed T1w for reconall
abcd_prefreesurfer_prep:
run: Off
# Will run Freesurfer for surface-based analysis. Will output traditional Freesurfer derivatives.
# If you wish to employ Freesurfer outputs for brain masking or tissue segmentation in the voxel-based pipeline,
# select those 'Freesurfer-' labeled options further below in anatomical_preproc.
freesurfer:
run_reconall: Off
# Ingress freesurfer recon-all folder
ingress_reconall: Off
# Add extra arguments to recon-all command
reconall_args:
# Run ABCD-HCP post FreeSurfer and fMRISurface pipeline
post_freesurfer:
run: Off
subcortical_gray_labels: /opt/dcan-tools/pipeline/global/config/FreeSurferSubcorticalLabelTableLut.txt
freesurfer_labels: /opt/dcan-tools/pipeline/global/config/FreeSurferAllLut.txt
surf_atlas_dir: /opt/dcan-tools/pipeline/global/templates/standard_mesh_atlases
gray_ordinates_dir: /opt/dcan-tools/pipeline/global/templates/Greyordinates
gray_ordinates_res: 2
high_res_mesh: 164
low_res_mesh: 32
fmri_res: 2
smooth_fwhm: 2
surface_connectivity:
run: Off
surface_parcellation_template: /cpac_templates/Schaefer2018_200Parcels_17Networks_order.dlabel.nii
longitudinal_template_generation:
# If you have multiple T1w's, you can generate your own run-specific custom
# T1w template to serve as an intermediate to the standard template for
# anatomical registration.
# This runs before the main pipeline as it requires multiple T1w sessions
# at once.
run: Off
# Freesurfer longitudinal template algorithm using FSL FLIRT
# Method to average the dataset at each iteration of the template creation
# Options: median, mean or std
average_method: median
# Degree of freedom for FLIRT in the template creation
# Options: 12 (affine), 9 (traditional), 7 (global rescale) or 6 (rigid body)
dof: 12
# Interpolation parameter for FLIRT in the template creation
# Options: trilinear, nearestneighbour, sinc or spline
interp: trilinear
# Cost function for FLIRT in the template creation
# Options: corratio, mutualinfo, normmi, normcorr, leastsq, labeldiff or bbr
cost: corratio
# Number of threads used for one run of the template generation algorithm
thread_pool: 2
# Threshold of transformation distance to consider that the loop converged
# (-1 means numpy.finfo(np.float64).eps and is the default)
convergence_threshold: -1
References¶
Cao, Q., Zang, Y., Sun, L., Sui, M., Long, X., Zou, Q., Wang, Y., 2006. Abnormal neural activity in children with attention deficit hyperactivity disorder: a resting-state functional magnetic resonance imaging study. Neuroreport 17, 1033-1036.
He, Y., Wang, L., Zang, Y., Tian, L., Zhang, X., Li, K., Jiang, T., 2007. Regional coherence changes in the early stages of Alzheimer’s disease: a combined structural and resting-state functional MRI study. Neuroimage 35, 488-500.
Kendall, M.G., Gibbons, J.D., 1990. Rank Correlation Methods, 5th ed. Oxford University Press.
Liu, H., Liu, Z., Liang, M., Hao, Y., Tan, L., Kuang, F., Yi, Y., Xu, L., Jiang, T., 2006. Decreased regional homogeneity in schizophrenia: a resting state functional magnetic resonance imaging study. Neuroreport 17, 19-22.
Wu, T., Zang, Y., Wang, L., Long, X., Hallett, M., Chen, Y., Li, K., Chan, P., 2007. Normal aging decreases regional homogeneity of the motor areas in the resting state. Neurosci Lett 422, 164-168.
Xiang-Yu Long, Xi-Nian Zuo, Vesa Kiviniemi, Yihong Yang, Qi-Hong Zou, Chao-Zhe Zhu, Tian-Zi Jiang, Hong Yang, Qi-Yong Gong, LiangWang, Kun-Cheng Li, Sheng Xie, Yu-Feng Zang. Default mode network as revealed with multiple methods for resting-state functional MRI analysis, J Neurosci Methods (2008), 171(2):349-55
Yan, C.-G., & Zang, Y.-F. (2010). DPARSF: A MATLAB Toolbox for “Pipeline” Data Analysis of Resting-State fMRI. Frontiers in systems neuroscience, 4, 13. doi:10.3389/fnsys.2010.00013
Zang, Y., Jiang, T., Lu, Y., He, Y., Tian, L., 2004. Regional homogeneity approach to fMRI data analysis. Neuroimage 22, 394-400.