Check Your Outputs¶
C-PAC ≥ 1.8.0¶
The output structure of C-PAC ≥ 1.8.0 is based on the Brain Imaging Data Structure v1.6.0-dev BIDS Derivatives specification. The output directory structure is a simple tree like
Changed in version 1.8.5.
└── pipeline-name
├── log
│ └── pipeline_{pipeline-name}
│ └── sub-{sub}_ses-{ses}
├── output
│ └── pipeline_{pipeline-name}
│ └── sub-{sub}
│ └── ses-{ses}
│ ├── anat
│ └── func
└── working
└── pipeline_{pipeline-name}
└── cpac_pipeline_{pipeline-name}_sub-{sub}_ses-{ses}
New in version 1.8.4.
Beginning with C-PAC v1.8.4, C-PAC generates a YAML file called sub-{sub}_ses-{ses}_expectedOutputs.yml near the beginning of each participant run. This file is stored in the participant’s log directory and lists derivative modalities (e.g., anat, func) and partial BIDS filenames for outputs expected to be generated and stored for each modality given the provided data and pipeline configuration. At the end of the run (or when crashing gracefully), C-PAC will check the output directory for the presence of all expected outputs. If any expected outputs are missing, C-PAC will generate a sub-{sub}_ses-{ses}_missingOutputs.yml file listing those missing outputs, also in the participant’s log directory.
Note
If any nodes use more memory at runtime than C-PAC estimates, C-PAC will report those instances near the end of the log in the terminal and in a log file called callback.log.resource_overusage.txt (beginning in v1.8.0). Please report excessive memory usage to the C-PAC team.
C-PAC ≤ 1.7.2¶
A standard C-PAC run output directory for one participant-session is shown below. A directory like this will exist for each participant-session in the run.
This is with all derivatives enabled, and the output, smoothing, and z-scoring options all at “Default”:
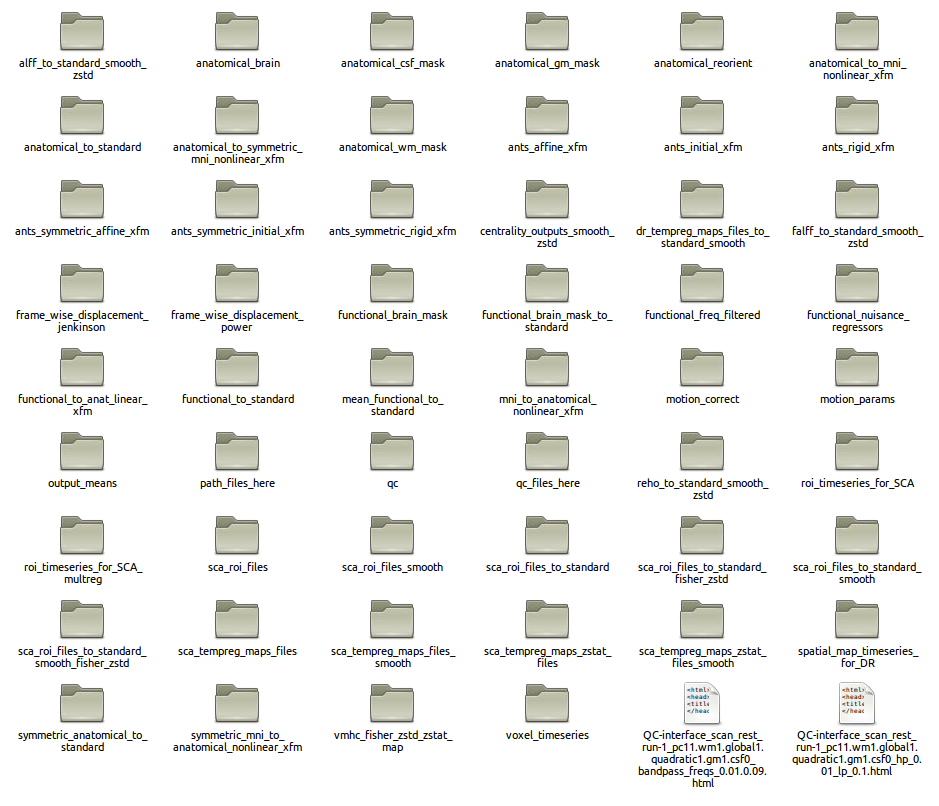
Descriptions¶
The output directory folders below are produced during a default run. If you enabled the additional outputs and have the full directory, proceed to the “Full Directory” section further below for descriptions of these extra outputs.
alff_to_standard_smooth_zstd: Warped to standard template, smoothed, z-score standardized output of ALFF.
anatomical_brain: Skull-stripped brain in anatomical space. This is the direct output of skull-stripping.
anatomical_csf_mask: Binary mask of the CSF in anatomical space. This is a result of anatomical segmentation.
anatomical_gm_mask: Binary mask of the gray matter in anatomical space. This is a result of anatomical segmentation.
anatomical_reorient: Deobliqued, reoriented whole-head anatomical scan. No other pre-processing yet.
anatomical_to_mni_nonlinear_xfm: Nonlinear warp transform from anatomical space to template space. Either ANTS or FSL-FNIRT warp depending on which was used.
anatomical_to_standard: Anatomical whole-head scan warped to standard/template.
anatomical_to_symmetric_mni_nonlinear_xfm: Present only if VMHC is run- nonlinear warp transform from anatomical space to symmetric template space. Either ANTS or FSL-FNIRT warp depending on which was used.
anatomical_wm_mask: Binary mask of the white matter in anatomical space. This is a result of anatomical segmentation.
ants_affine_xfm: ANTS only- linear affine warp from anatomical space to template space.
ants_initial_xfm: ANTS only- “initial” linear warp from anatomical space to template space.
ants_rigid_xfm: ANTS only- rigid linear warp from anatomical space to template space.
ants_symmetric_affine_xfm: ANTS only- Same as above, but the warp to symmetric template space.
ants_symmetric_initial_xfm: ANTS only- Same as above, but the warp to symmetric template space.
ants_symmetric_rigid_xfm: ANTS only- Same as above, but the warp to symmetric template space.
centrality_outputs_smooth_zstd: Smoothed, z-score standardized centrality outputs (already in template space). Sub-directories in this folder for each type of centrality that was run (Degree, Eigenvector, LFCD).
dr_tempreg_maps_files_to_standard_smooth: Warped to standard template, smoothed Dual Regression outputs. Sub-directories in this folder for each map provided.
falff_to_standard_smooth_zstd: Warped to standard template, smoothed, z-score standardized output of f/ALFF.
frame_wise_displacement_jenkinson: 1-D file containing the vector of framewise displacement values between volumes, as calculated via Jenkinson.
frame_wise_displacement_power: 1-D file containing the vector of framewise displacement values between volumes, as calculated via Power.
functional_brain_mask: Binary mask of the brain in functional space.
functional_brain_mask_to_standard: Binary mask of the functional-space brain warped to standard template.
functional_freq_filtered: Preprocessed functional timeseries file all the way up to temporal filtering. 4D time series.
functional_nuisance_regressors: .mat file containing the data corresponding to each nuisance that was regressed out during nuisance regression.
functional_to_anat_linear_xfm: Functional-to-anatomical space linear transform. FSL-FLIRT format.
functional_to_standard: Preprocessed functional timeseries warped to standard template space. 4D time series.
functional_to_standard_smooth: Smoothed version of functional_to-standard. 4D time series.
functional_to_standard_xfm: Composite transform (as a NIfTI .nii.gz file) bringing data from native functional (BOLD) space to template space.
mean_functional_to_standard: Mean functional (one-volume 3D file of functional scan) warped to standard template space.
mni_to_anatomical_nonlinear_xfm: Same as the anatomical_to_mni_nonlinear_xfm described above, except the inverse warp.
motion_correct: Motion-corrected functional timeseries in functional space, before the rest of functional preprocessing. 4D time series.
motion_params: Text file containing the single-value max or mean numbers of each head motion parameter/measure.
output_means: Text files containing the mean intensity values of each output or derivative. Used later in group-level analysis.
path_files_here: Text files containing full file paths to all of the C-PAC outputs in the output directory. Can be used for convenient file path parsing.
qc: PNG image files of all QC Interface montages, graphs, and charts. Can be easily viewed in one place in the QC-interface_{scan}_{nuisance/preprocessing strategy}.html files in the output directory (described below).
qc_files_here: Individual QC Interface HTML pages. Used later in the main QC Interface index generation.
reho_to_standard_smooth_zstd: Warped to standard template, smoothed, z-score standardized output of Regional Homogeneity (ReHo).
roi_timeseries_for_SCA: CSV files containing the extracted ROI timeseries for each ROI provided for Seed-Based Correlation Analysis (SCA).
roi_timeseries_for_SCA_multreg: Same as above, but the extracted ROI timeseries for each ROI provided for Multiple Regression (if different from SCA).
sca_roi_files: Raw correlation outputs of Seed-Based Correlation Analysis (SCA) for each ROI provided. Sub-directories in this folder for each ROI.
sca_roi_files_smooth: Smoothed version of the above.
sca_roi_files_to_standard: Warped to standard template version of sca_roi_files.
sca_roi_files_to_standard_fisher_zstd: Fisher r-to-z transformed version of sca_roi_files_to_standard.
sca_roi_files_to_standard_smooth: Smoothed version of sca_roi_files_to_standard.
sca_roi_files_to_standard_smooth_fisher_zstd: Fisher r-to-z transformed version of sca_roi_files_to_standard_smooth.
sca_tempreg_maps_files: Multiple Regression output files (already in template space).
sca_tempreg_maps_files_smooth: Smoothed version of sca_tempreg_maps_files.
sca_tempreg_maps_zstat_files: Z-stat file outputs of Multiple Regression. Produced by the –out_z option of FSL’s fsl_glm tool.
sca_tempreg_maps_zstat_files_smooth: Smoothed version of sca_tempreg_maps_zstat_files.
spatial_map_timeseries_for_DR: Text file containing the GLM output of the timeseries associated with the voxels in the spatial map provided to Spatial Regression. Used later in Temporal Regression to complete the Dual Regression derivative.
symmetric_anatomical_to_standard: Same as anatomical_to_standard, except warped to the symmetric anatomical template instead.
symmetric_mni_to_anatomical_nonlinear_xfm: Same as mni_to_anatomical_nonlinear_xfm, except the inverse warp from the symmetric template back to anatomical space.
vmhc_fisher_zstd_zstat_map: Fisher r-to-z transformed, Z-stat output of Voxel-Mirrored Homotopic Connectivity (VMHC).
voxel_timeseries: 1-D file (and also CSV and .npz files, if selected) containing the voxel-wise extracted timeseries based on each ROI provided. Sub-directories in this folder for each ROI.
QC-interface_{scan}_{nuisance/preprocessing strategy}.html: QC Interface HTML page for each scan and nuisance/preprocessing strategy combination.
Sub-Directories¶
Each folder in the output directory may have a different amount of sub-directories, depending on how many functional scans specified in the data configuration, or how many pipeline customizations and forked strategies you specified in the pipeline configuration. For example, if there are multiple functional scans, you’ll see a folder for each one if you enter any of the functional-derived outputs’ folders, as seen below:
An example of multiple sub-directories for multiple nuisance regression strategies:
And sub-directories for each ROI provided for a derivative that may take in multiple ROIs from a mask or atlas- for example, Seed-Based Correlation Analysis (SCA) in this case:
Full Directory¶
The C-PAC output directory is considerably larger when “Extra Functional Outputs”, “Debugging Outputs”, and both Smoothed/Non-smoothed and both z-score standardized/raw outputs are all enabled.
In addition to the output directories described above under “Descriptions”, the following outputs are also written to the output directory when all of the output options mentioned above are enabled:
Debugging Outputs - Set ‘Write Debugging Outputs’ to ‘On’ to produce these outputs.
coordinate_transformation: Output of the AFNI 3dvolreg -1Dmatrix_save flag, when run during functional pre-processing. This is the matrix transformation of the base to input DICOM coordinates.
dr_tempreg_maps_zstat_files_to_standard: Warped to standard statistical Z-stat outputs of FSL GLM (run during dual regression), via the –out_z flag.
dr_tempreg_maps_zstat_files_to_standard_smooth: Same as above, but smoothed.
max_displacement: Output of the AFNI 3dvolreg -maxdisp1D flag, when run during functional pre-processing. This is a 1D file containing the maximum displacement (in mm) for each volume.
movement_parameters: 1D file containing six movement/motion parameters (3 Translation, 3 Rotations) in different columns (roll pitch yaw dS dL dP), as output by AFNI 3dVolreg.
power_params: Text file containing the power parameters resulting from the calculation of Mean Framewise Displacement.
vmhc_fisher_zstd: The Fisher’s r-to-z transformed output of VMHC, before calculation of the Z-statistic.
vmhc_raw_score: The direct, ‘raw’ output of VMHC, before performing Fisher’s r-to-z, and before calculating the Z-statistic.
Extra Functional Outputs - Set ‘Write Extra Functional Outputs’ to ‘On’ to produce these outputs.
functional_nuisance_residuals: A NIfTI (.nii) file of the pre-processed functional time series produced directly after nuisance regression is performed. 4D time series.
functional_nuisance_residuals_smooth: Smoothed version of functional_nuisance_residuals. 4D time series.
functional_preprocessed: The functional time series produced directly after initial functional pre-processing (de-obliquing, re-orienting, motion correction, functional skull-stripping, and image intensity normalization). In native space. 4D time series.
functional_preprocessed_mask: A binary mask of the functional_preprocessed output. In native space.
mean_functional: The mean of the functional time-series taken over the time course. Presented as a single-volume NifTI file.
mean_functional_in_anat: The mean of the functional time-series, registered/warped to anatomical (T1) space.
motion_correct_to_standard: Motion-corrected functional timeseries in template space, before the rest of functional preprocessing. 4D time series.
motion_correct_to_standard_smooth: Motion-corrected functional timeseries in template space, before the rest of functional preprocessing, but smoothed. 4D time series.
slice_time_corrected: The functional time-series after slice-time correction. 4D time series.
Non-smoothed - Set ‘Run Smoothing’ to either ‘Off’ or ‘On/Off’ to produce these outputs.
alff_to_standard_zstd: Warped-to-standard, z-scored output of ALFF, without smoothing.
dr_tempreg_maps_files_to_standard: Warped to standard, non-smoothed Dual Regression outputs. Sub-directories in this folder for each map provided.
falff_to_standard_zstd: Warped to standard, z-scored outputs of f/ALFF, without smoothing.
reho_to_standard_zstd: Warped to standard, z-scored outputs of ReHo, without smoothing.
Raw scores (before z-scoring) - Set ‘z-score Standardize Derivatives’ to either ‘Off’ or ‘On/Off’ to produce these outputs.
alff_to_standard_smooth: Warped-to-standard, smoothed output of ALFF, without z-scoring.
falff_to_standard_smooth: Warped to standard, smoothed output of f/ALFF, without z-scoring.
reho_to_standard_smooth: Warped to standard, smoothed output of ReHo, without z-scoring.
Both non-smoothed and raw scores - Both ‘Run Smoothing’ and ‘z-score Standardize Derivatives’ set to either ‘Off’ or ‘On/Off’.
alff_to_standard: Warped to standard output of ALFF, without smoothing and without z-scoring.
falff_to_standard: Warped to standard output of f/ALFF, without smoothing and without z-scoring.
reho_to_standard: Warped to standard output of ReHo, without smoothing and without z-scoring.
Native space (not warped to standard) - Set ‘Run Functional to Template Registration’ to ‘On/Off’ to produce these outputs.
alff: The direct output of ALFF, before warping to standard space.
falff: The direct output of f/ALFF, before warping to standard space.
reho: The direct output of ReHo, before warping to standard space.
Visual Quality Control¶
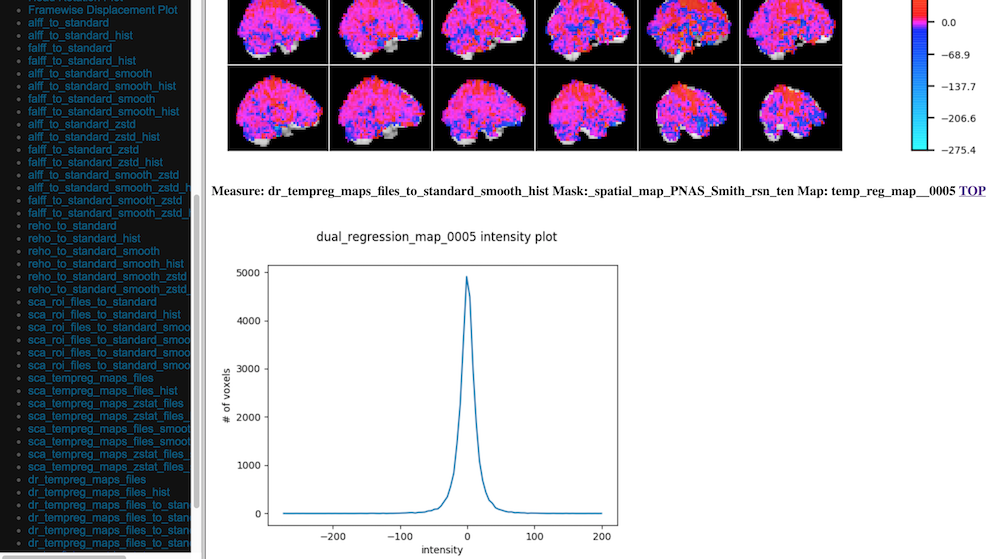
C-PAC’s data quality control (QC) interface allows you to take a quick glance at the overall quality of your results (registration quality, signal-to-noise ratio, movement plots, computed derivative histograms, etc.). In its current form, the QC interface is a collection of HTML pages - one for each participant-scan-nuisance regression strategy combination, and they can be found in the Output Directory under each participant’s directory level.
Upcoming Additions
In future releases, more visualizations will be introduced, and the QC pages will be integrated into an easy-to-use dashboard that is updated throughout the process of your C-PAC run, and also provides information on the status of the pipeline. As always, we greatly appreciate any ideas, suggestions, or items on your wishlist and take them into consideration.
Quick Look¶
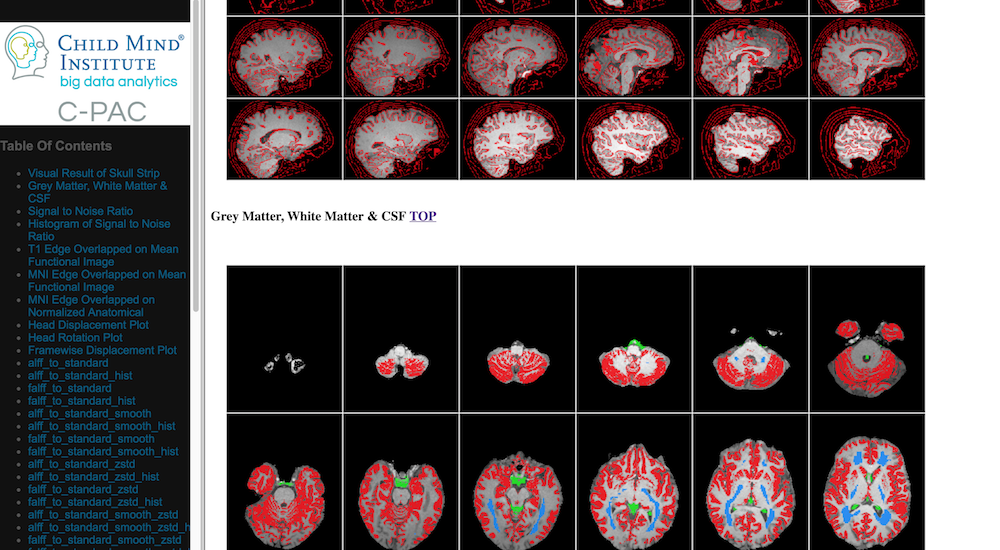
Skull-stripping and Segmentation Quality
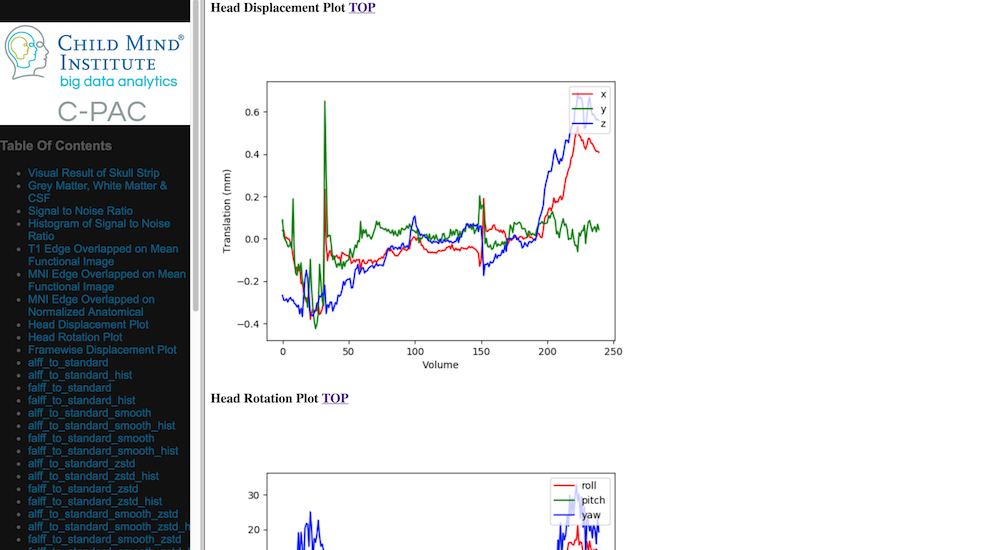
Head Movement Measures
Quick View of Derivatives
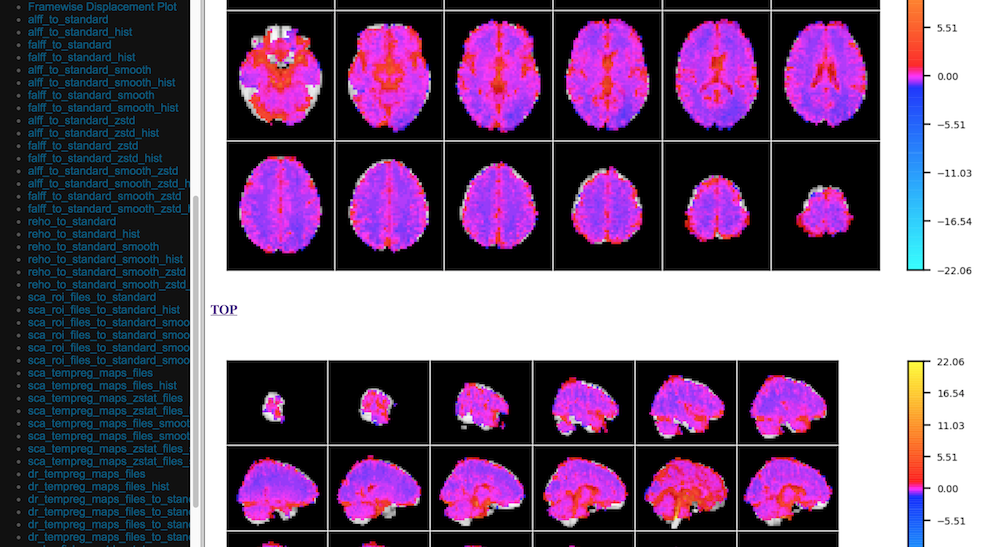
With Histograms