Voxel-Mirrored Homotopic Connectivity (VMHC)¶
Introduction & Background¶
Functional homotopy, the synchrony in patterns of spontaneous activity between homotopic (geometrically corresponding) regions in each hemisphere, is a fundamental characteristic of the brain’s functional architecture (Salvador et al., 2005). Though homotopic patterns of resting functional connectivity have been observed across the brain, the strength of this connectivity can vary between regions (Stark et al., 2008). This variation (seen in the figure below taken from Stark et al., 2008) is thought to reflect hemispheric and regional specialization in information processing.
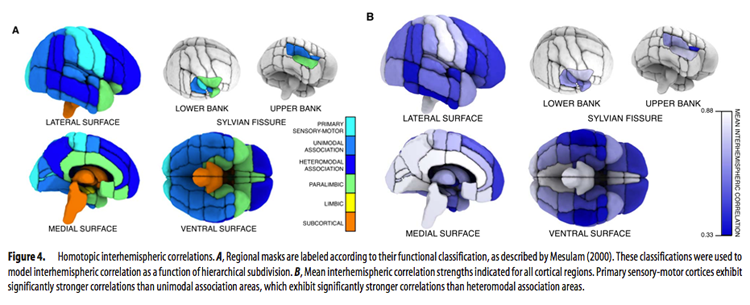
Voxel-Mirrored Homotopic Connectivity (VMHC) quantifies functional homotopy by providing a voxel-wise measure of connectivity between hemispheres. This is done by computing the connectivity between each voxel in one hemisphere and its mirrored counterpart in the other (Zuo et al., 2010).
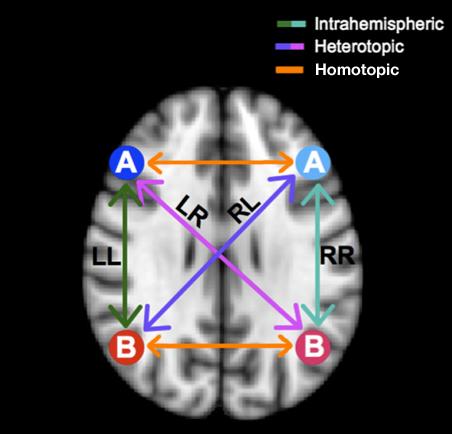
The schematic above (adapted from Gee et al., 2011) illustrates how VMHC compares mirrored voxels across hemispheres in a symmetrical brain.
Computation and Analysis Considerations¶
VMHC assumes symmetric morphology between hemispheres. As this assumption does not hold for real brains, images must be transformed before VMHC can be calculated. In CPAC, anatomical images are transformed to fit a symmetric template. Functional data is then transformed to fit the new symmetrical anatomical image. VMHC is computed as the functional connectivity between any pair of symmetric interhemispheric voxels. For more details on how CPAC computes these steps, please see the VMHC Page of the developer documentation.
It is worth noting that VMHC is somewhat affected by the level of spatial smoothing used. Specifically, smoothing improves the correspondence between homotopic regions and results in greater VMHC strength across subjects. Importantly however, smoothing does not affect the overall pattern of VMHC.
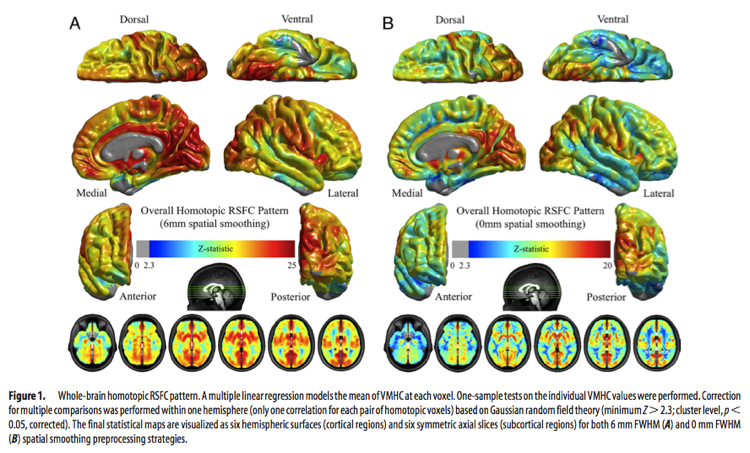
The image above (taken from Zuo et al., 2010) shows differences in VMHC due to smoothing. These differences should be taken into account when comparing studies using VMHC.
Applications and Recommendations¶
Despite being a relatively new analysis method, a number of studies have already utilized VMHC to investigate group differences in functional homotopy. Kelly et al. (2011) found reduced VMHC between prefrontal regions in patients with cocaine addiction compared to healthy controls, and Zuo and colleagues (2010) were able to chart changing functional homotopy across healthy aging.
Other recent studies examining functional homotopy (but not using VMHC) have found reduced interhemispheric connectivity in patients with autism (Anderson et al., 2011), and that negative correlations between hemispheres were stronger than positive correlations within a hemisphere (Gee et al. 2001).
Future studies may benefit from using VMHC as an indicator of disturbed functional specialization in patient groups, as well as in longitudinal studies to investigate changes in functional homotopy over time due to treatment, intervention, or development.
Configuring CPAC to run VMHC¶
Voxel-Mirrored Homotopic Connectivity (VMHC) - [On, Off]: Calculate Voxel-Mirrored Homotopic Connectivity (VMHC) for all voxels.
Symmetric Brain Template - [path]: Included as part of the ‘Image Resource Files’ package available on the Install page of the User Guide. It is not necessary to change this path unless you intend to use a non-standard symmetric template.
Symmetric Brain + Skull Template - [path]: Included as part of the ‘Image Resource Files’ package available on the Install page of the User Guide. It is not necessary to change this path unless you intend to use a non-standard symmetric template.
Dilated Symmetric Brain Mask - [path]: Included as part of the ‘Image Resource Files’ package available on the Install page of the User Guide. It is not necessary to change this path unless you intend to use a non-standard symmetric template.
FLIRT Configuration File - [path]: Included as part of the ‘Image Resource Files’ package available on the Install page of the User Guide. It is not necessary to change this path unless you intend to use a non-standard symmetric template.
Configuration Without the GUI¶
The following nested key/value pairs will be set to these defaults if not defined in your pipeline configuration YAML.
voxel_mirrored_homotopic_connectivity:
# VMHC
# Calculate Voxel-mirrored Homotopic Connectivity (VMHC) for all voxels.
run: On
symmetric_registration:
# Included as part of the 'Image Resource Files' package available on the Install page of the User Guide.
# It is not necessary to change this path unless you intend to use a non-standard symmetric template.
T1w_brain_template_symmetric: $FSLDIR/data/standard/MNI152_T1_${resolution_for_anat}_brain_symmetric.nii.gz
# Included as part of the 'Image Resource Files' package available on the Install page of the User Guide.
# It is not necessary to change this path unless you intend to use a non-standard symmetric template.
T1w_brain_template_symmetric_funcreg: $FSLDIR/data/standard/MNI152_T1_${func_resolution}_brain_symmetric.nii.gz
# A reference symmetric brain template for resampling
T1w_brain_template_symmetric_for_resample: $FSLDIR/data/standard/MNI152_T1_1mm_brain_symmetric.nii.gz
# Included as part of the 'Image Resource Files' package available on the Install page of the User Guide.
# It is not necessary to change this path unless you intend to use a non-standard symmetric template.
T1w_template_symmetric: $FSLDIR/data/standard/MNI152_T1_${resolution_for_anat}_symmetric.nii.gz
# Included as part of the 'Image Resource Files' package available on the Install page of the User Guide.
# It is not necessary to change this path unless you intend to use a non-standard symmetric template.
T1w_template_symmetric_funcreg: $FSLDIR/data/standard/MNI152_T1_${func_resolution}_symmetric.nii.gz
# A reference symmetric skull template for resampling
T1w_template_symmetric_for_resample: $FSLDIR/data/standard/MNI152_T1_1mm_symmetric.nii.gz
# Included as part of the 'Image Resource Files' package available on the Install page of the User Guide.
# It is not necessary to change this path unless you intend to use a non-standard symmetric template.
dilated_symmetric_brain_mask: $FSLDIR/data/standard/MNI152_T1_${resolution_for_anat}_brain_mask_symmetric_dil.nii.gz
# A reference symmetric brain mask template for resampling
dilated_symmetric_brain_mask_for_resample: $FSLDIR/data/standard/MNI152_T1_1mm_brain_mask_symmetric_dil.nii.gz
References¶
Gee, D. G., Biswal, B. B., Kelly, C., Stark, D. E., Margulies, D. S., Shehzad, Z., Uddin, L. Q., et al. (2011). Low frequency fluctuations reveal integrated and segregated processing among the cerebral hemispheres. Neuroimage, 54(1), 517–527.
Kelly, C., Zuo, X.-N., Gotimer, K., Cox, C. L., Lynch, L., Brock, D., Imperati, D., et al. (2011). Reduced interhemispheric resting state functional connectivity in cocaine addiction. Biological Psychiatry, 69(7), 684–692.
Salvador R, Suckling J, Coleman MR, Pickard JD, Menon D, Bullmore E (2005) Neurophysiological architecture of functional magnetic resonance images of human brain. Cereb Cortex 15:1332–1342.
Stark, D. E., Margulies, D. S., Shehzad, Z. E., Reiss, P., Kelly, A. M. C., Uddin, L. Q., Gee, D. G., et al. (2008). Regional variation in interhemispheric coordination of intrinsic hemodynamic fluctuations. The Journal of Neuroscience, 28(51), 13754–13764.
Zuo, X.-N., Kelly, C., Di Martino, A., Mennes, M., Margulies, D. S., Bangaru, S., Grzadzinski, R., et al. (2010). Growing together and growing apart: regional and sex differences in the lifespan developmental trajectories of functional homotopy. The Journal of Neuroscience, 30(45), 15034–15043.