ICA-AROMA (Independent Component Analysis - Automatic Removal of Motion Artifacts)¶
INTRODUCTION¶
Motion artifacts, when induced in fMRI can cause true effects to be classified into noise, or correlate motion artifacts to true effects.While most methods for dealing with the artifacts may result in reduction of temporal freedom or destroy autocorrelation structure, and such drawbacks can be overcome by using AROMA-ICA(). ICA-AROMA concerns a data-driven method to identify and remove motion-related independent components from fMRI data.To that end it exploits a small, but robust set of theoretically motivated features, preventing the need for classifier re-training and therefore providing direct and easy applicability.
COMPUTATION AND ANALYSIS CONSIDERATION:¶
This method uses an ICA-based strategy for Automatic Removal of Motion Artifacts (ICA-AROMA) that uses a small (n=4), but robust set of theoretically motivated temporal and spatial features. Our strategy does not require classifier re-training, retains the data’s autocorrelation structure and largely preserves temporal degrees of freedom.ICA-AROMA identified motion components with high accuracy and robustness as illustrated by leave-N-out cross-validation.(Pruim R1 et.al.,NeuroImage,2015). C-PAC’s implementation of Aroma-ICA involves using the nipype’s interface to the package developed in FSL (https://github.com/maartenmennes/ICA-AROMA). The output directory includes the denoised file along with the melodic directory, statistics and other feature classification texts. The movement parameters necessary for running this script is obtained using FSL’s McFLIRT, and the mask file is obtained using FSL’s BET (as per suggestion).
INSTALLING AROMA-ICA:¶
To run AROMA-ICA using C-PAC, it is essential to download and set up AROMA-ICA in your system. This can be accomplished by:
mkdir -p /opt/ICA-AROMA
curl -sSL "https://github.com/rhr-pruim/ICA-AROMA/archive/v0.4.3-beta.tar.gz" | tar -xzC /opt/ICA-AROMA --strip-components 1
chmod +x /opt/ICA-AROMA/ICA_AROMA.py
CONFIGURING C-PAC TO RUN AROMA-ICA:¶
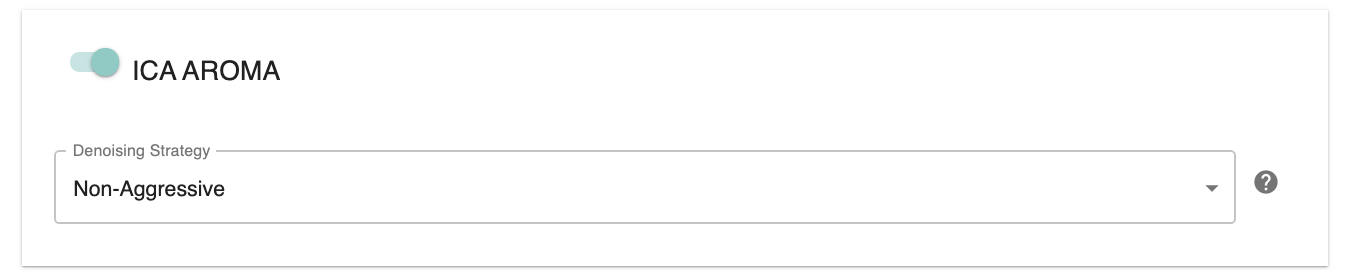
Run Aroma-ICA: Choose “On” to run Aroma-ICA.
Denoise type: Choose between ‘nonaggr’ for non-aggressively denoised file and ‘aggr’ aggressively denoised file.
CONFIGURING AROMA-ICA USING A YAML FILE:¶
The following nested key/value pairs that will be set to these defaults if not defined in your pipeline configuration YAML:
# - if 'single_step_resampling_from_stc', 'template' is the only valid option for ``nuisance_corrections: 2-nuisance_regression: space``
using: 'default'
functional_preproc:
run: On
update_header:
# Convert raw data from LPI to RPI
run: On
truncation:
# First timepoint to include in analysis.
# Default is 0 (beginning of timeseries).
# First timepoint selection in the scan parameters in the data configuration file, if present, will over-ride this selection.
# Note: the selection here applies to all scans of all participants.
start_tr: 0
# Last timepoint to include in analysis.
# Default is None or End (end of timeseries).
# Last timepoint selection in the scan parameters in the data configuration file, if present, will over-ride this selection.
# Note: the selection here applies to all scans of all participants.
stop_tr: None
scaling:
# Scale functional raw data, usually used in rodent pipeline
run: Off
# Scale the size of the dataset voxels by the factor.
scaling_factor: 10
despiking:
# Run AFNI 3dDespike
# this is a fork point
# run: [On, Off] - this will run both and fork the pipeline
run: [Off]
slice_timing_correction:
# Interpolate voxel time courses so they are sampled at the same time points.
# this is a fork point
# run: [On, Off] - this will run both and fork the pipeline
run: [On]
# use specified slice time pattern rather than one in header
tpattern: None
# align each slice to given time offset
# The default alignment time is the average of the 'tpattern' values (either from the dataset header or from the tpattern option).
tzero: None
motion_estimates_and_correction:
run: On
motion_estimates:
# calculate motion statistics BEFORE slice-timing correction
calculate_motion_first: Off
# calculate motion statistics AFTER motion correction
calculate_motion_after: On
motion_correction:
# using: ['3dvolreg', 'mcflirt']
# Forking is currently broken for this option.
# Please use separate configs if you want to use each of 3dvolreg and mcflirt.
# Follow https://github.com/FCP-INDI/C-PAC/issues/1935 to see when this issue is resolved.
using: ['3dvolreg']
# option parameters
AFNI-3dvolreg:
# This option is useful when aligning high-resolution datasets that may need more alignment than a few voxels.
functional_volreg_twopass: On
# Choose motion correction reference. Options: mean, median, selected_volume, fmriprep_reference
motion_correction_reference: ['mean']
# Choose motion correction reference volume
motion_correction_reference_volume: 0
motion_estimate_filter:
# Filter physiological (respiration) artifacts from the head motion estimates.
# Adapted from DCAN Labs filter.
# https://www.ohsu.edu/school-of-medicine/developmental-cognition-and-neuroimaging-lab
# https://www.biorxiv.org/content/10.1101/337360v1.full.pdf
# this is a fork point
# run: [On, Off] - this will run both and fork the pipeline
run: [Off]
filters:
- # options: "notch", "lowpass"
filter_type: "notch"
# Number of filter coefficients.
filter_order: 4
# Dataset-wide respiratory rate data from breathing belt.
# Notch filter requires either:
# "breathing_rate_min" and "breathing_rate_max"
# or
# "center_frequency" and "filter_bandwitdh".
# Lowpass filter requires either:
# "breathing_rate_min"
# or
# "lowpass_cutoff".
# If "breathing_rate_min" (for lowpass and notch filter)
# and "breathing_rate_max" (for notch filter) are set,
# the values set in "lowpass_cutoff" (for lowpass filter),
# "center_frequency" and "filter_bandwidth" (for notch filter)
# options are ignored.
# Lowest Breaths-Per-Minute in dataset.
# For both notch and lowpass filters.
breathing_rate_min:
# Highest Breaths-Per-Minute in dataset.
# For notch filter.
breathing_rate_max:
# notch filter direct customization parameters
# mutually exclusive with breathing_rate options above.
# If breathing_rate_min and breathing_rate_max are provided,
# the following parameters will be ignored.
# the center frequency of the notch filter
center_frequency:
# the width of the notch filter
filter_bandwidth:
- # options: "notch", "lowpass"
filter_type: "lowpass"
# Number of filter coefficients.
filter_order: 4
# Dataset-wide respiratory rate data from breathing belt.
# Notch filter requires either:
# "breathing_rate_min" and "breathing_rate_max"
# or
# "center_frequency" and "filter_bandwitdh".
# Lowpass filter requires either:
# "breathing_rate_min"
# or
# "lowpass_cutoff".
# If "breathing_rate_min" (for lowpass and notch filter)
# and "breathing_rate_max" (for notch filter) are set,
# the values set in "lowpass_cutoff" (for lowpass filter),
# "center_frequency" and "filter_bandwidth" (for notch filter)
# options are ignored.
# Lowest Breaths-Per-Minute in dataset.
# For both notch and lowpass filters.
breathing_rate_min:
# lowpass filter direct customization parameter
# mutually exclusive with breathing_rate options above.
# If breathing_rate_min is provided, the following
# parameter will be ignored.
# the frequency cutoff of the filter
lowpass_cutoff:
distortion_correction:
# this is a fork point
# run: [On, Off] - this will run both and fork the pipeline
run: [On]
# using: ['PhaseDiff', 'Blip', 'Blip-FSL-TOPUP']
# PhaseDiff - Perform field map correction using a single phase difference image, a subtraction of the two phase images from each echo. Default scanner for this method is SIEMENS.
# Blip - Uses AFNI 3dQWarp to calculate the distortion unwarp for EPI field maps of opposite/same phase encoding direction.
# Blip-FSL-TOPUP - Uses FSL TOPUP to calculate the distortion unwarp for EPI field maps of opposite/same phase encoding direction.
using: ['PhaseDiff', 'Blip']
# option parameters
PhaseDiff:
# Since the quality of the distortion heavily relies on the skull-stripping step, we provide a choice of method ('AFNI' for AFNI 3dSkullStrip or 'BET' for FSL BET).
# Options: 'BET' or 'AFNI'
fmap_skullstrip_option: 'BET'
# Set the fraction value for the skull-stripping of the magnitude file. Depending on the data, a tighter extraction may be necessary in order to prevent noisy voxels from interfering with preparing the field map.
# The default value is 0.5.
fmap_skullstrip_BET_frac: 0.5
# Set the threshold value for the skull-stripping of the magnitude file. Depending on the data, a tighter extraction may be necessary in order to prevent noisy voxels from interfering with preparing the field map.
# The default value is 0.6.
fmap_skullstrip_AFNI_threshold: 0.6
Blip-FSL-TOPUP:
# (approximate) resolution (in mm) of warp basis for the different sub-sampling levels, default 10
warpres: 10
# sub-sampling scheme, default 1
subsamp: 1
# FWHM (in mm) of gaussian smoothing kernel, default 8
fwhm: 8
# Max # of non-linear iterations, default 5
miter: 5
# Weight of regularisation, default depending on --ssqlambda and --regmod switches. See user documentation.
lambda: 1
# If set (=1), lambda is weighted by current ssq, default 1
ssqlambda: 1
# Model for regularisation of warp-field [membrane_energy bending_energy], default bending_energy
regmod: bending_energy
# Estimate movements if set, default 1 (true)
estmov: 1
# Minimisation method 0=Levenberg-Marquardt, 1=Scaled Conjugate Gradient, default 0 (LM)
minmet: 0
# Order of spline, 2->Qadratic spline, 3->Cubic spline. Default=3
splineorder: 3
# Precision for representing Hessian, double or float. Default double
numprec: double
# Image interpolation model, linear or spline. Default spline
interp: spline
# If set (=1), the images are individually scaled to a common mean, default 0 (false)
scale: 0
# If set (=1), the calculations are done in a different grid, default 1 (true)
regrid: 1
func_masking:
run: On
# using: ['AFNI', 'FSL', 'FSL_AFNI', 'Anatomical_Refined', 'Anatomical_Based', 'Anatomical_Resampled', 'CCS_Anatomical_Refined']
# FSL_AFNI: fMRIPrep-style BOLD mask. Ref: https://github.com/nipreps/niworkflows/blob/a221f612/niworkflows/func/util.py#L246-L514
# Anatomical_Refined: 1. binarize anat mask, in case it is not a binary mask. 2. fill holes of anat mask 3. init_bold_mask : input raw func → dilate init func brain mask 4. refined_bold_mask : input motion corrected func → dilate anatomical mask 5. get final func mask
# Anatomical_Based: Generate the BOLD mask by basing it off of the anatomical brain mask. Adapted from DCAN Lab's BOLD mask method from the ABCD pipeline.
# Anatomical_Resampled: Resample anatomical brain mask in standard space to get BOLD brain mask in standard space. Adapted from DCAN Lab's BOLD mask method from the ABCD pipeline. ("Create fMRI resolution standard space files for T1w image, wmparc, and brain mask […] don't use FLIRT to do spline interpolation with -applyisoxfm for the 2mm and 1mm cases because it doesn't know the peculiarities of the MNI template FOVs")
# CCS_Anatomical_Refined: Generate the BOLD mask by basing it off of the anatomical brain. Adapted from the BOLD mask method from the CCS pipeline.
# this is a fork point
using: ['AFNI']
FSL-BET:
# Apply to 4D FMRI data, if bold_bet_functional_mean_boolean : Off.
# Mutually exclusive with functional, reduce_bias, robust, padding, remove_eyes, surfaces
# It must be 'on' if select 'reduce_bias', 'robust', 'padding', 'remove_eyes', or 'bet_surfaces' on
functional_mean_boolean: Off
# Set an intensity threshold to improve skull stripping performances of FSL BET on rodent scans.
functional_mean_thr:
run: Off
threshold_value: 98
# Bias correct the functional mean image to improve skull stripping performances of FSL BET on rodent scans
functional_mean_bias_correction: Off
# Set the threshold value controling the brain vs non-brain voxels.
frac: 0.3
# Mesh created along with skull stripping
mesh_boolean: Off
# Create a surface outline image
outline: Off
# Add padding to the end of the image, improving BET.Mutually exclusive with functional,reduce_bias,robust,padding,remove_eyes,surfaces
padding: Off
# Integer value of head radius
radius: 0
# Reduce bias and cleanup neck. Mutually exclusive with functional,reduce_bias,robust,padding,remove_eyes,surfaces
reduce_bias: Off
# Eyes and optic nerve cleanup. Mutually exclusive with functional,reduce_bias,robust,padding,remove_eyes,surfaces
remove_eyes: Off
# Robust brain center estimation. Mutually exclusive with functional,reduce_bias,robust,padding,remove_eyes,surfaces
robust: Off
# Create a skull image
skull: Off
# Gets additional skull and scalp surfaces by running bet2 and betsurf. This is mutually exclusive with reduce_bias, robust, padding, remove_eyes
surfaces: Off
# Apply thresholding to segmented brain image and mask
threshold: Off
# Vertical gradient in fractional intensity threshold (-1,1)
vertical_gradient: 0.0
FSL_AFNI:
bold_ref:
brain_mask: $FSLDIR/data/standard/MNI152_T1_${resolution_for_anat}_brain_mask.nii.gz
brain_probseg: $FSLDIR/data/standard/MNI152_T1_${resolution_for_anat}_brain_mask.nii.gz
Anatomical_Refined:
# Choose whether or not to dilate the anatomical mask if you choose 'Anatomical_Refined' as the functional masking option. It will dilate one voxel if enabled.
anatomical_mask_dilation: False
# Apply functional mask in native space
apply_func_mask_in_native_space: On
generate_func_mean:
# Generate mean functional image
run: On
normalize_func:
# Normalize functional image
run: On
coreg_prep:
# Generate sbref
run: On
nuisance_corrections:
1-ICA-AROMA:
# this is a fork point
# run: [On, Off] - this will run both and fork the pipeline
run: [Off]
# Types of denoising strategy:
# nonaggr: nonaggressive-partial component regression
# aggr: aggressive denoising
denoising_type: nonaggr
EXTERNAL RESOURCES:¶
AROMA-ICA python script: https://github.com/maartenmennes/ICA-AROMA
REFERENCES:¶
Neuroimage. 2015 May 15;112:267-277. doi: 10.1016/j.neuroimage.2015.02.064. Epub 2015 Mar 11.
https://github.com/maartenmennes/ICA-AROMA/blob/master/Manual.pdf